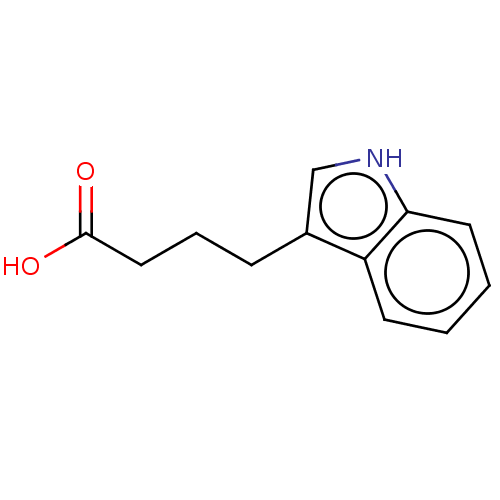
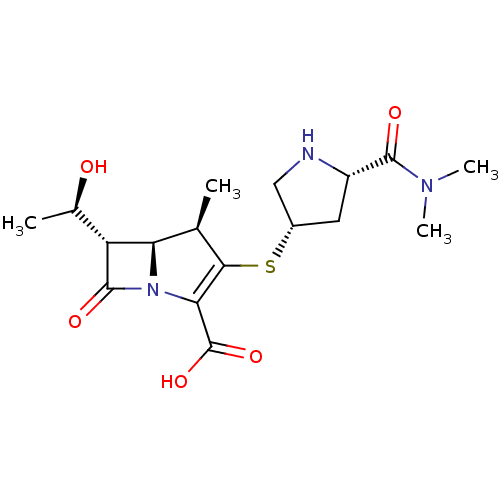
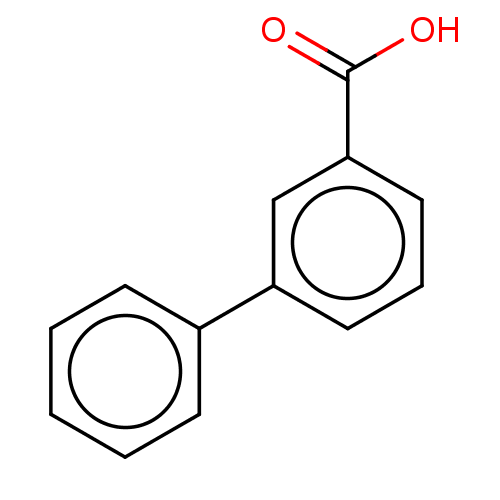
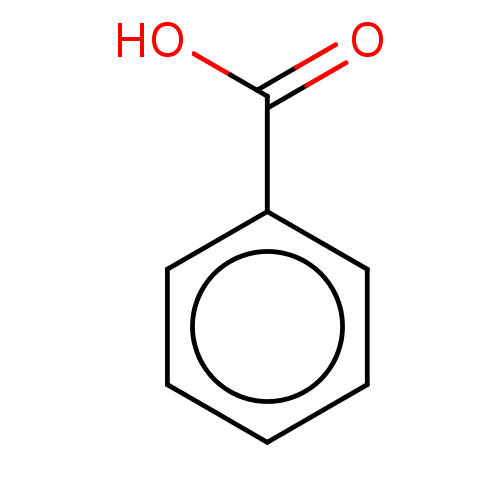
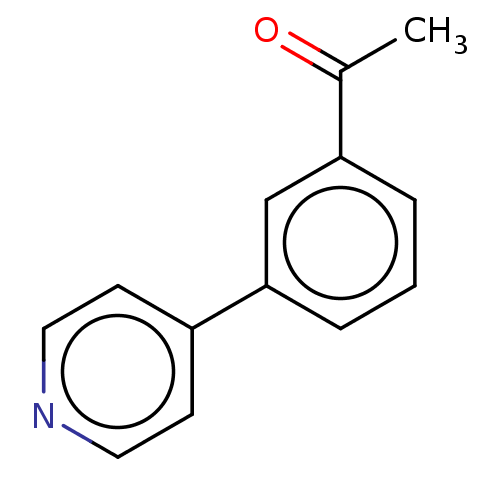
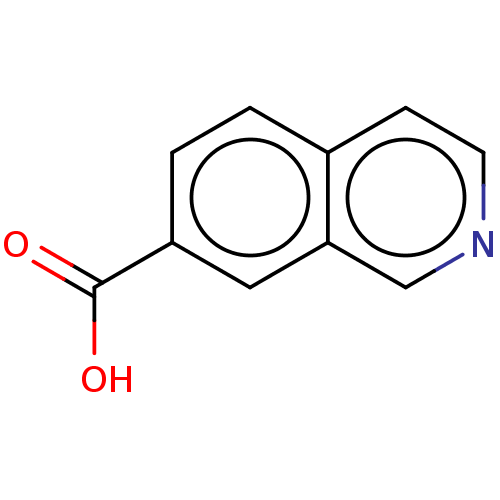
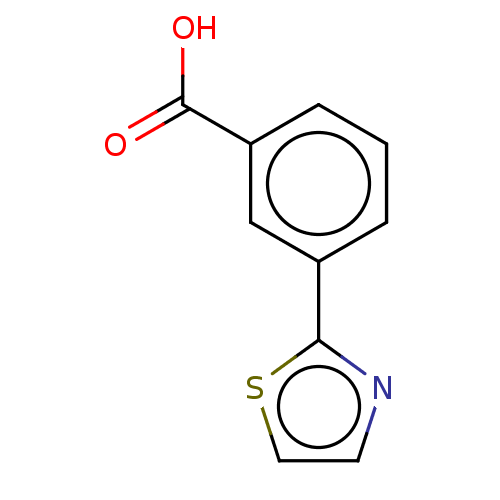
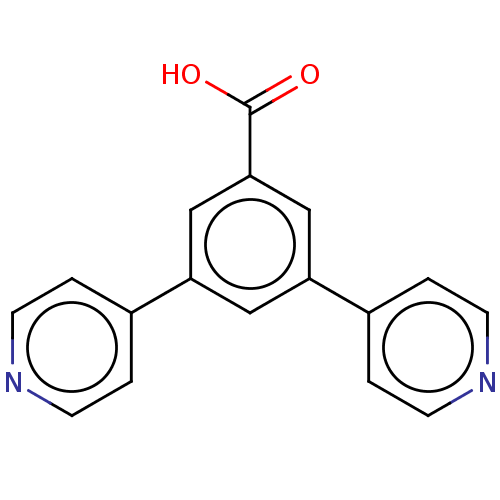
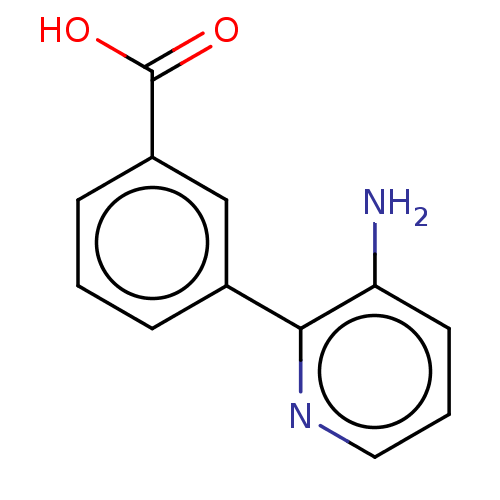
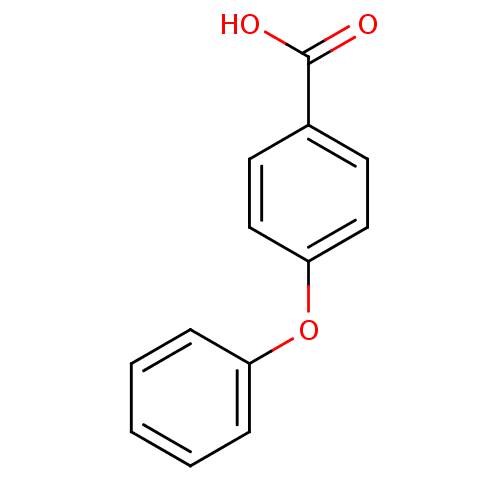
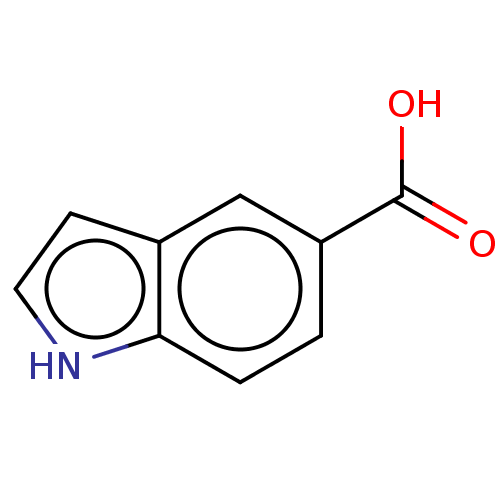
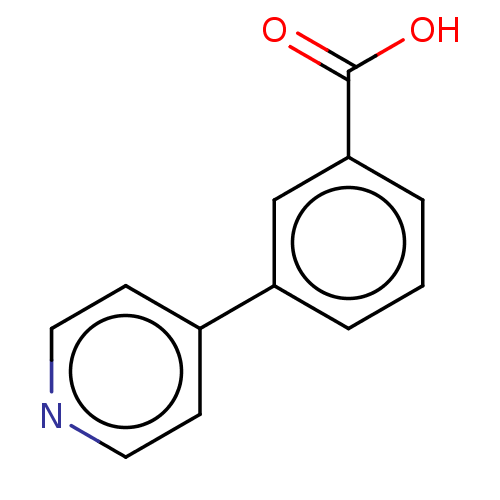
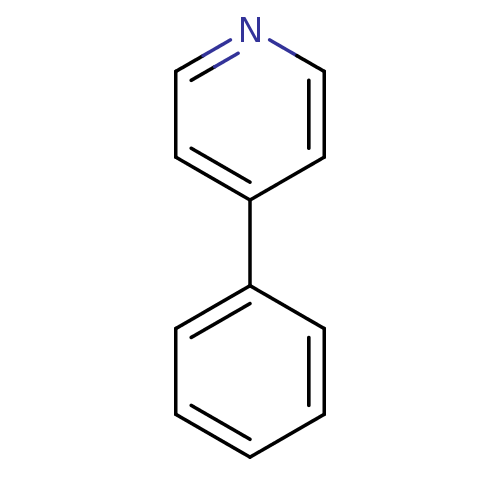
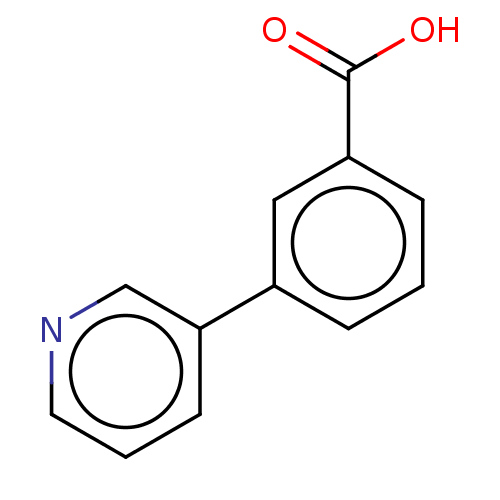
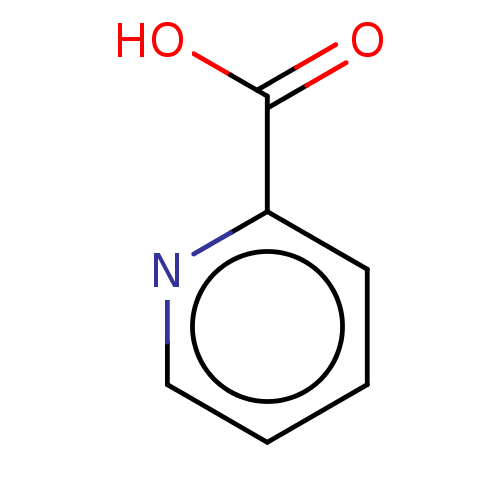
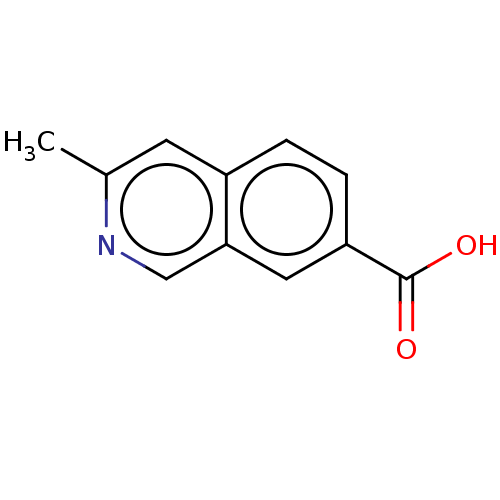
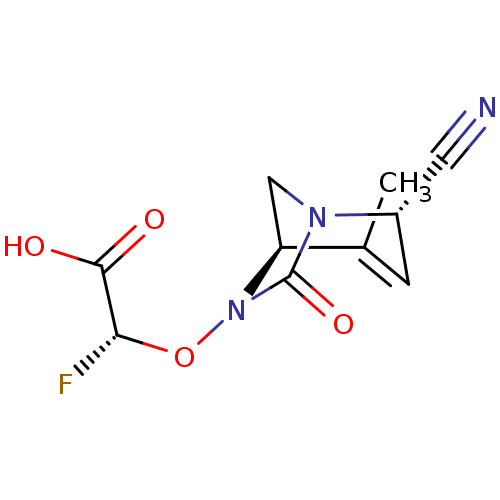
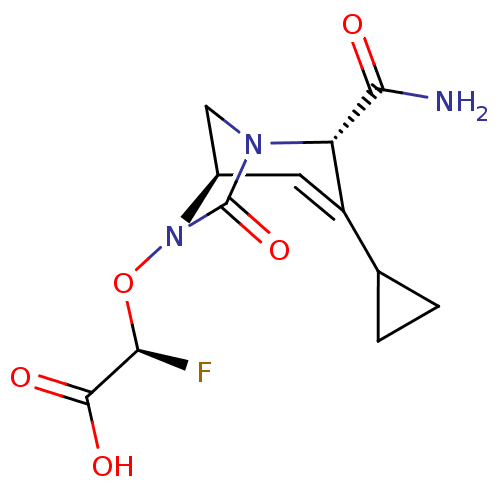
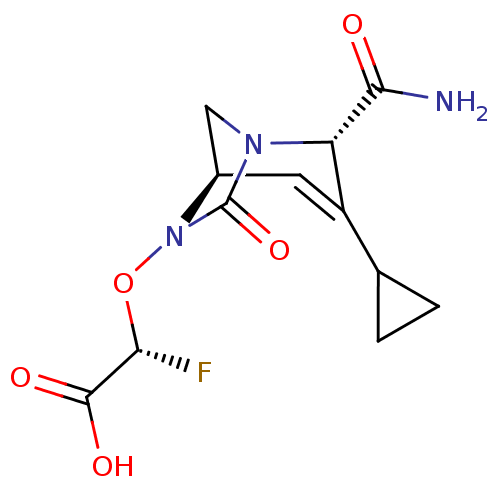
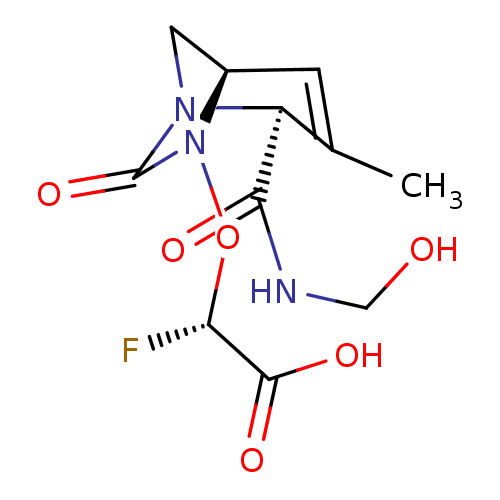
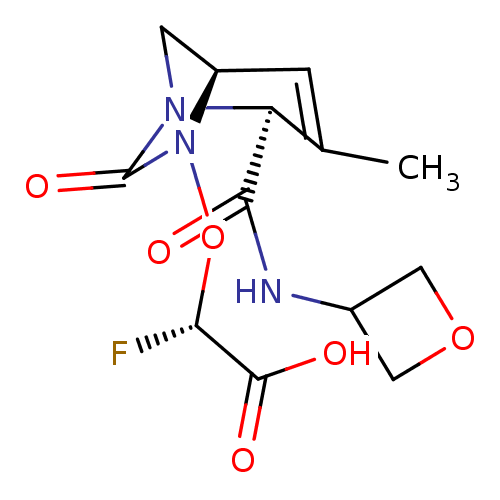
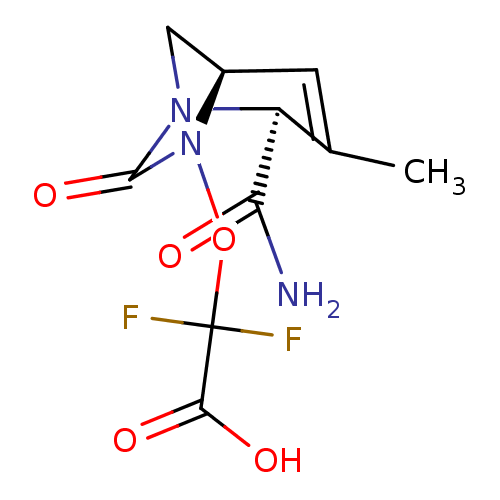
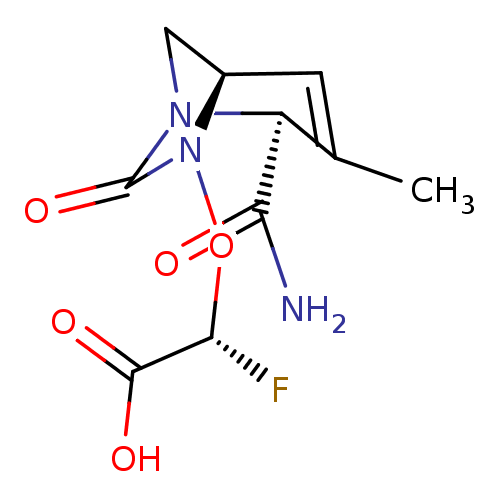
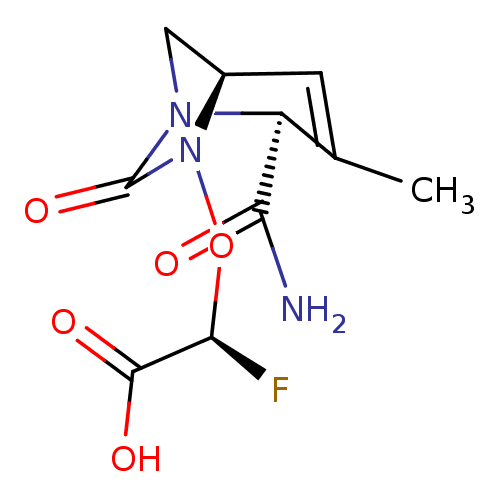
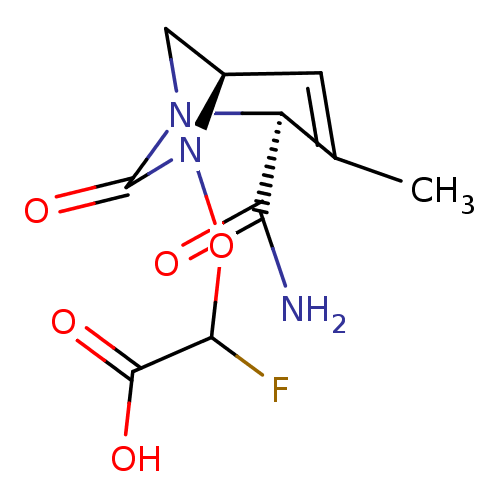
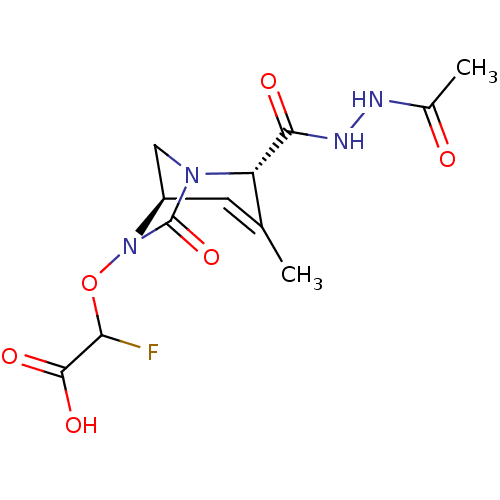
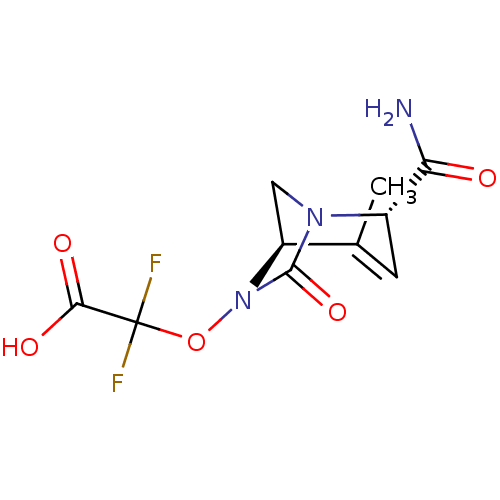
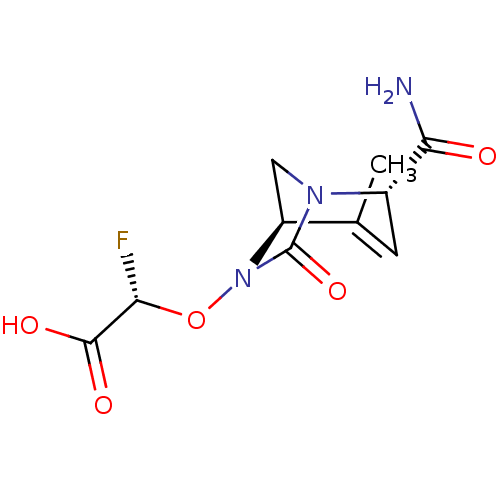
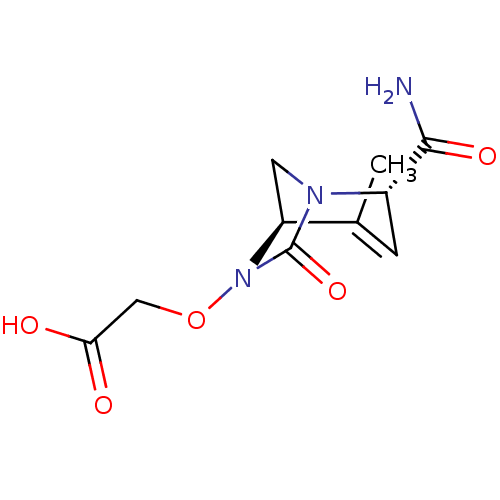
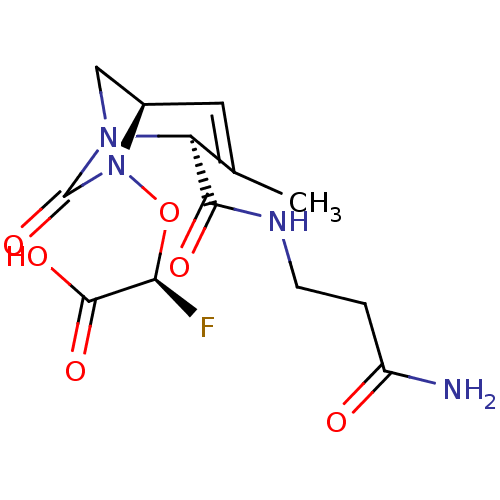
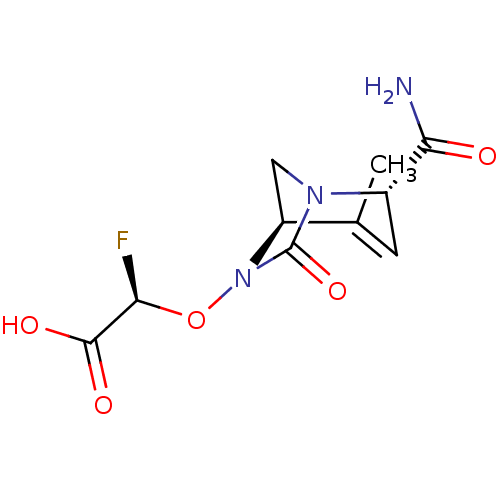
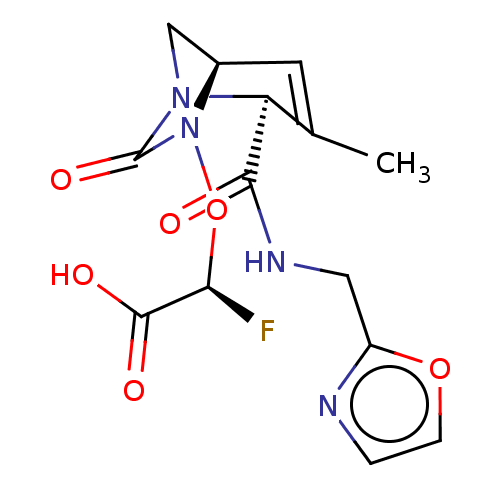
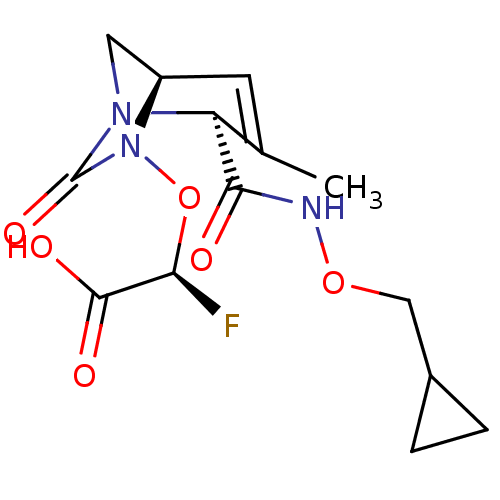
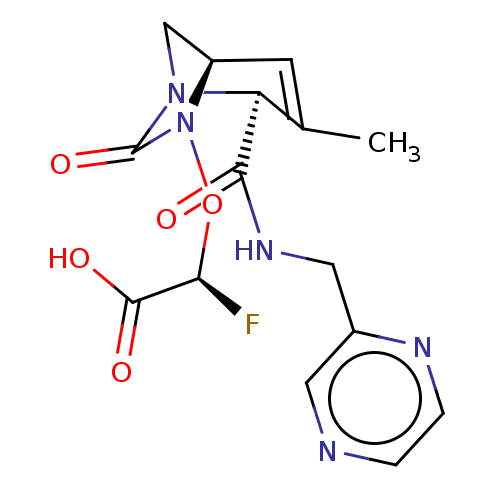
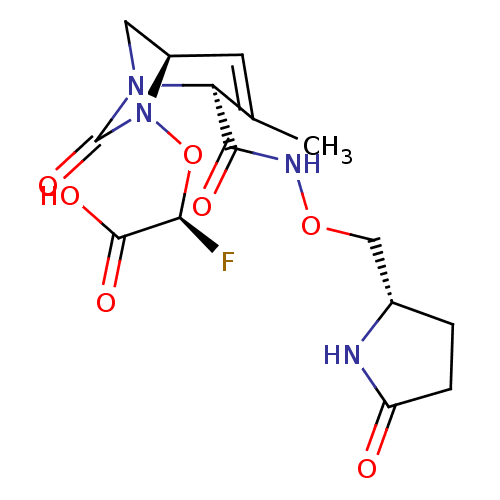
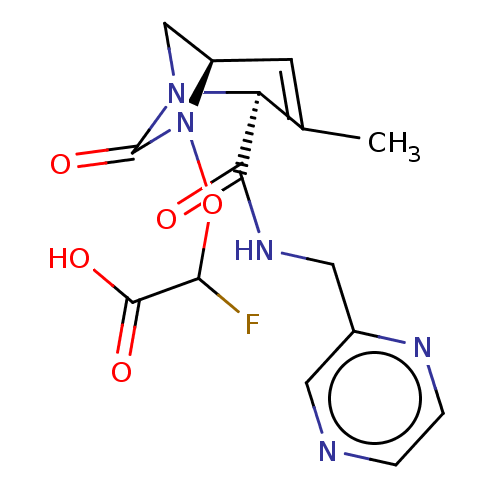
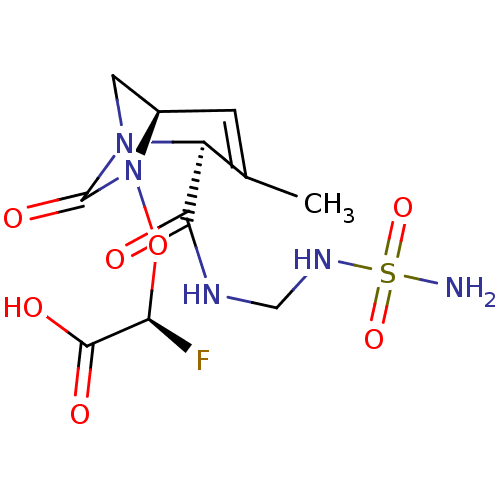
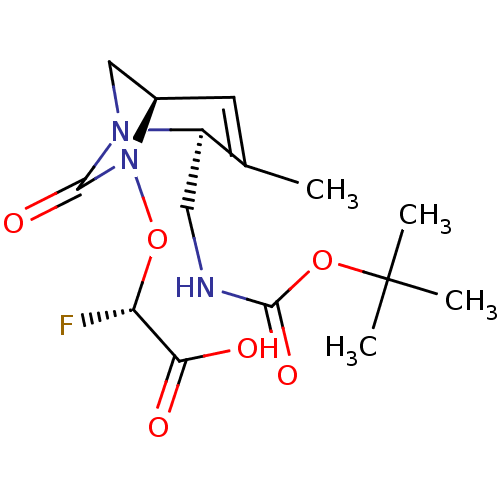
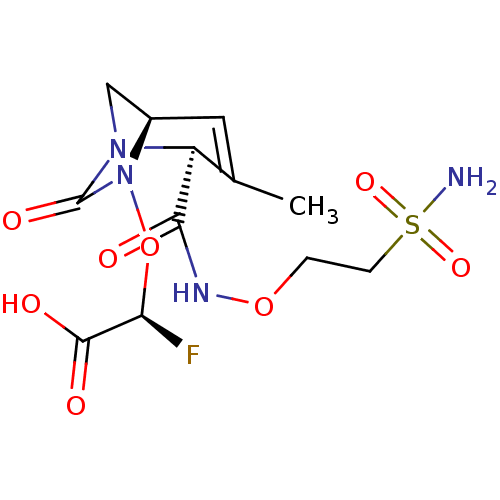
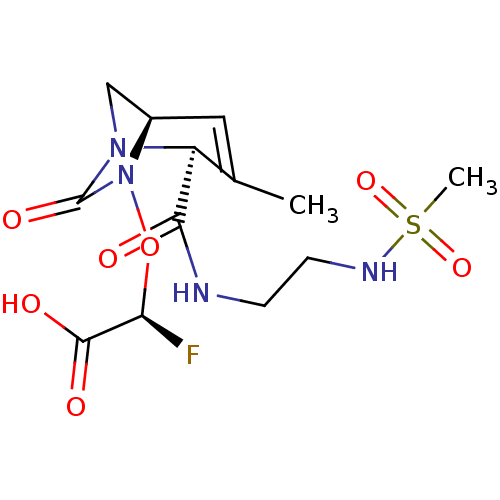
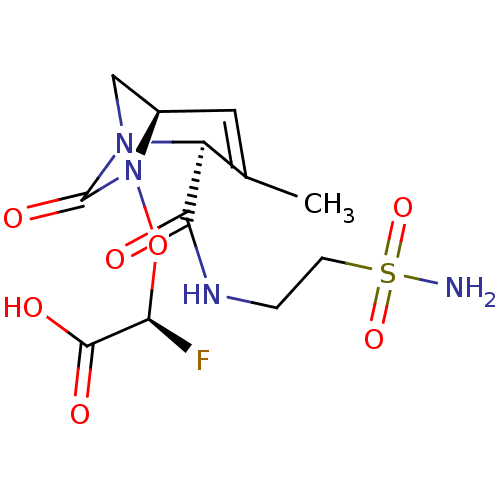
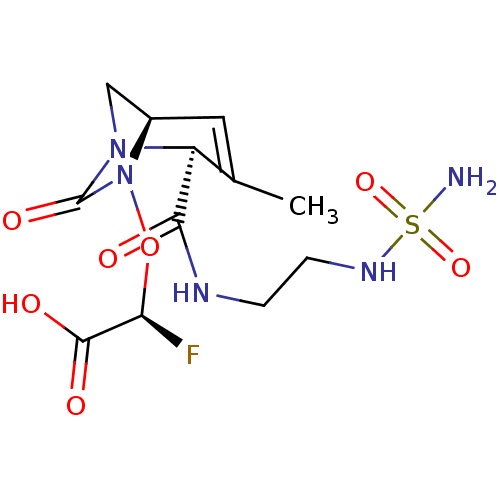
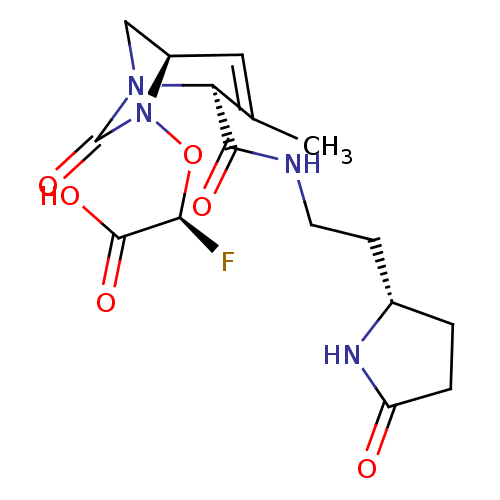
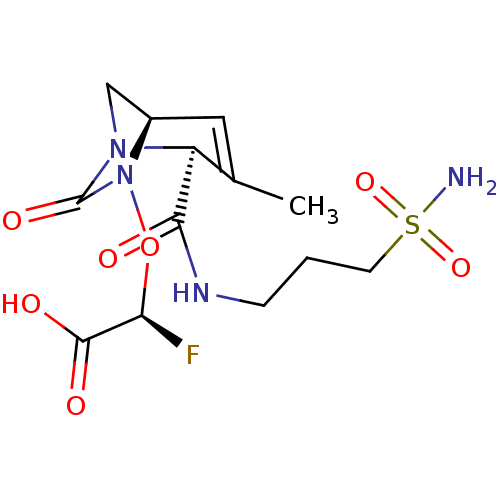
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 4.30E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
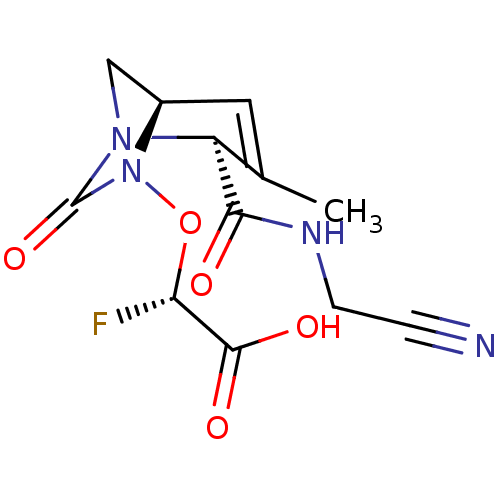
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 850nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
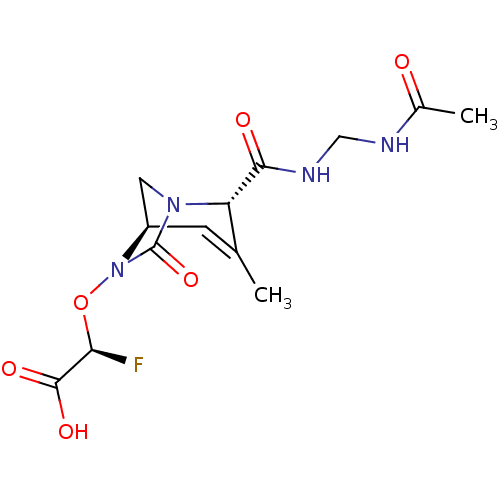
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 3.70E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
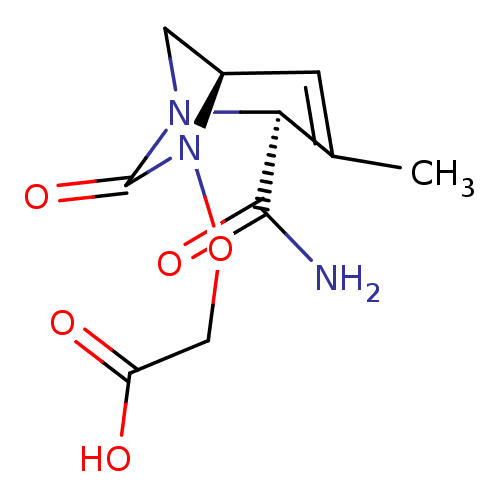
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: >1.00E+6nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: >1.00E+6nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 1.18E+6nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 3.10E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 5.00E+4nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 3.30E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 6.60E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 1.38E+6nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 2.80E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKon: 0.346M-1s-1Assay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: >1.00E+6nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 2.80E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: >1.00E+6nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataKd: 7.00E+5nMAssay Description:Binding affinity to native signal deficient and TEV cleavage site containing His-tagged Klebsiella pneumoniae OXA-48 expressed in Escherichia coli as...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 1.30nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 5.20nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 6.40nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 7.30nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 11nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 12nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 14nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 15nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 15nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 17nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 19nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 28nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 45nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 51nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 53nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 75nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 77nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 78nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 87nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 110nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 110nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 140nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 150nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 220nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 220nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 230nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 340nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 410nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 420nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 530nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
TargetBeta-lactamase(Pseudomonas aeruginosa (g-Proteobacteria))
Uit The Arctic University of Norway
Curated by ChEMBL
Uit The Arctic University of Norway
Curated by ChEMBL
Affinity DataIC50: 590nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair