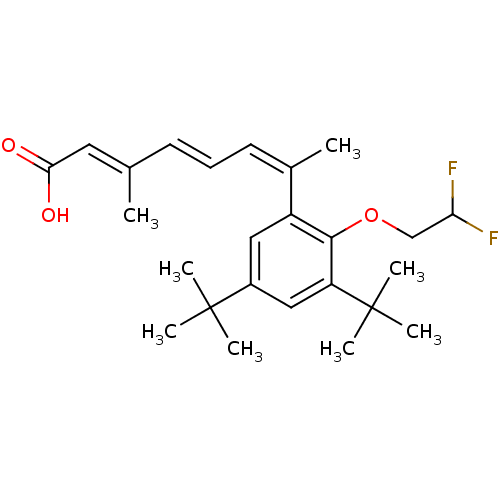
BDBM50129720 (2E,4E,6Z)-7-[3,5-Di-tert-butyl-2-(2,2-difluoro-ethoxy)-phenyl]-3-methyl-octa-2,4,6-trienoic acid::7-[3,5-Di-tert-butyl-2-(2,2-difluoro-ethoxy)-phenyl]-3-methyl-octa-2,4,6-trienoic acid::CHEMBL42314
SMILES C\C(\C=C\C=C(\C)c1cc(cc(c1OCC(F)F)C(C)(C)C)C(C)(C)C)=C/C(O)=O
InChI Key InChIKey=BHIBZAZKKARFIM-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 30 hits for monomerid = 50129720
Found 30 hits for monomerid = 50129720
Affinity DataKi: 1.5nMAssay Description:In vitro agonistic activity against RXR alpha in CV-1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 2.70nMAssay Description:Inhibition of 3[H]9-cis-retinoic acid binding to Retinoic acid receptor RXR-alpha expressed in CV-1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 3nMAssay Description:Binding affinity against Retinoic acid receptor RXR-alpha was determined in vitro by using [3H]9-cis-RA as radioligandMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma/Retinoic acid receptor RXR-alpha(Human)
Ligand Pharmaceuticals
Curated by ChEMBL
Ligand Pharmaceuticals
Curated by ChEMBL
Affinity DataEC50: 3nMAssay Description:Transcriptional activity against RXR:PPAR-gamma synergy was determined in vitroMore data for this Ligand-Target Pair
Affinity DataKi: 3nMAssay Description:Binding affinity for Retinoic acid receptor RXR-alpha was determined by competing with 3[H]-9-cis-RAMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma/Retinoic acid receptor RXR-alpha(Human)
Ligand Pharmaceuticals
Curated by ChEMBL
Ligand Pharmaceuticals
Curated by ChEMBL
Affinity DataEC50: 3.10nMAssay Description:Synergistic activation of BRL-49653 mediated PPAR gamma and retinoid X receptor alpha expressed in CV-1 cell transcriptional activation assayMore data for this Ligand-Target Pair
Affinity DataKi: 4nMAssay Description:Displacement of [3H]9-cis-RA from RXR beta receptor in CV-1 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 8nMAssay Description:Inhibitory activity against Retinoic acid receptor RXR-alpha antagonist was determined in vitroMore data for this Ligand-Target Pair
Affinity DataIC50: 8nMAssay Description:In vitro transcriptional activation in CV-1 cells expressing RXR-alphaMore data for this Ligand-Target Pair
Affinity DataIC50: 8nMAssay Description:Antagonist activity for Retinoic X receptor alpha in CV1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 9nMAssay Description:Binding affinity for Retinoic acid receptor RXR-beta was determined by competing with 3[H]-9-cis-RAMore data for this Ligand-Target Pair
Affinity DataEC50: 11nMAssay Description:Transcriptional activation in CV-1 cells expressing Retinoic X receptor alphaMore data for this Ligand-Target Pair
Affinity DataKi: 12nMAssay Description:Binding affinity for Retinoic acid receptor RXR-gamma was determined by competing with 3[H]-9-cis-RAMore data for this Ligand-Target Pair
Affinity DataKi: 18nMAssay Description:Displacement of [3H]9-cis-RA from Retinoic X receptor betaMore data for this Ligand-Target Pair
Affinity DataKi: 18nMAssay Description:Binding affinity against Retinoic acid receptor RXR-alpha by [3H]9-cis-RA displacement.More data for this Ligand-Target Pair
Affinity DataKi: 39nMAssay Description:Displacement of [3H]9-cis-RA from retinoic X receptor gammaMore data for this Ligand-Target Pair
Affinity DataKi: 54nMAssay Description:Binding affinity against RAR alpha receptor using [3H]ATRA as radioligand in CV-1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 84nMAssay Description:Binding affinity against RXR gamma receptor using [3H]9-cis-RA as radioligand in CV-1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Binding affinity against retinoic acid receptor gamma by [3H]ATRA displacement.More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Binding affinity for Retinoic acid receptor gamma was determined by competing with 3[H]-ATRAMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Binding affinity against retinoic acid receptor alpha by [3H]ATRA displacement.More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Binding affinity against retinoic acid receptor beta by [3H]ATRA displacement.More data for this Ligand-Target Pair
Affinity DataKi: 2.75E+3nMAssay Description:Binding affinity for Retinoic acid receptor alpha was determined by competing with 3[H]-ATRAMore data for this Ligand-Target Pair
Affinity DataKi: 4.69E+3nMAssay Description:Binding affinity for Retinoic acid receptor beta was determined by competing with 3[H]-ATRAMore data for this Ligand-Target Pair
Affinity DataKi: 5.59E+3nMAssay Description:Binding affinity for peroxisome proliferator activated receptor gammaMore data for this Ligand-Target Pair
Affinity DataKi: >9.51E+3nMAssay Description:Displacement of [3H]ATRA from RAR gamma receptor in CV-1 cells; Not testedMore data for this Ligand-Target Pair
Affinity DataKi: 9.51E+3nMAssay Description:Binding affinity against RAR beta receptor using [3H]ATRA as radioligand in CV-1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nMAssay Description:Binding affinity for Peroxisome proliferator activated receptor alpha was determinedMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nMAssay Description:Inhibition of 3[H]ATRA binding to retinoid acid receptor gamma expressed in CV-1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.00E+4nMAssay Description:Binding affinity against Retinoic acid receptor gamma was determined in vitro by using [3H]ATRA as radioligandMore data for this Ligand-Target Pair