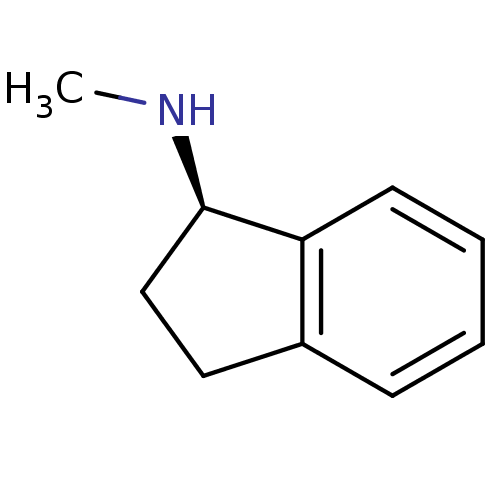
BDBM10995 (1R)-N-methyl-2,3-dihydro-1H-inden-1-amine::N-methyl-1(R)-aminoindan::R-MAI::rasagiline analog
SMILES CN[C@@H]1CCc2c1cccc2
InChI Key InChIKey=AIXUYZODYPPNAV-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 10995
Found 4 hits for monomerid = 10995
Affinity DataIC50: 200nMAssay Description:Inhibition of PDK1 (unknown origin) using PDKtide substrate and [gamma-32P]ATP by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:Inhibition of PDK1 (unknown origin) expressed in HEK293 cells using PDKtide as substrate after 10 mins by liquid scintillation counting in presence o...More data for this Ligand-Target Pair
Affinity DataKi: 1.70E+4nM ΔG°: -6.50kcal/molepH: 7.5 T: 2°CAssay Description:MAO A and MAO B activities were determined spectrophotometrically at 316 nm and 250 nm using kynuramine and benzylamine as substrates, respectively. ...More data for this Ligand-Target Pair
Affinity DataKi: 5.60E+4nM ΔG°: -5.80kcal/molepH: 7.5 T: 2°CAssay Description:MAO A and MAO B activities were determined spectrophotometrically at 316 nm and 250 nm using kynuramine and benzylamine as substrates, respectively. ...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)