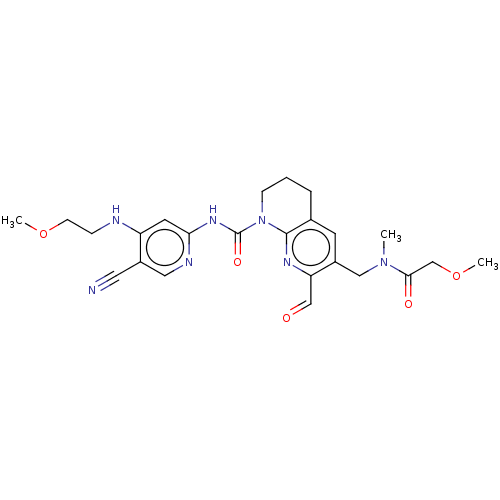
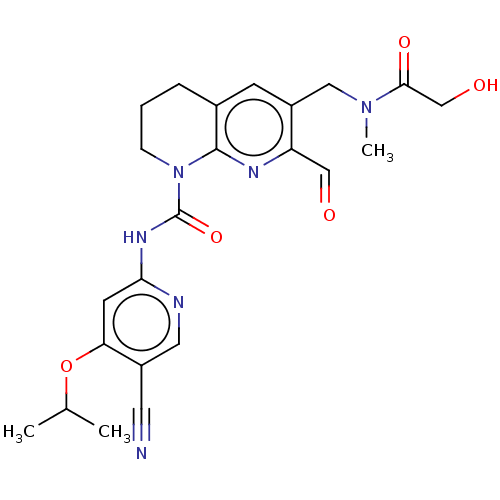
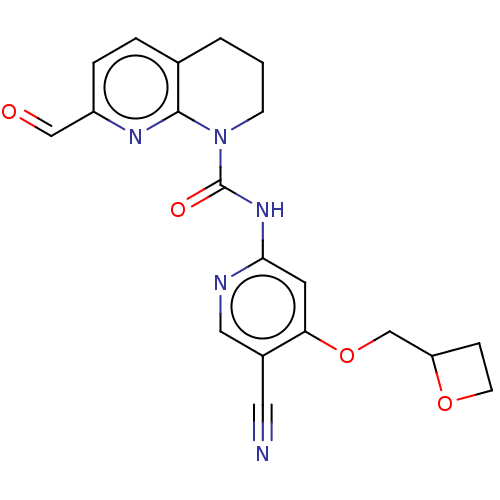
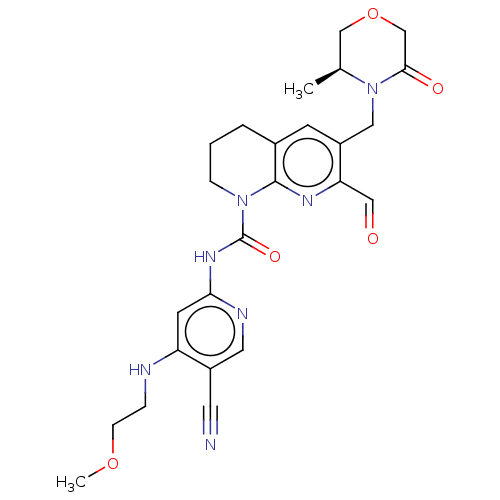
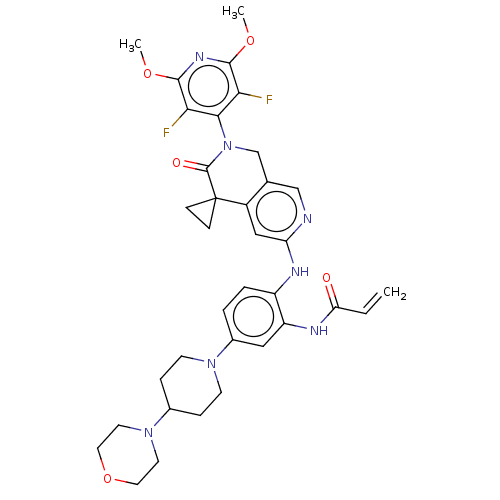
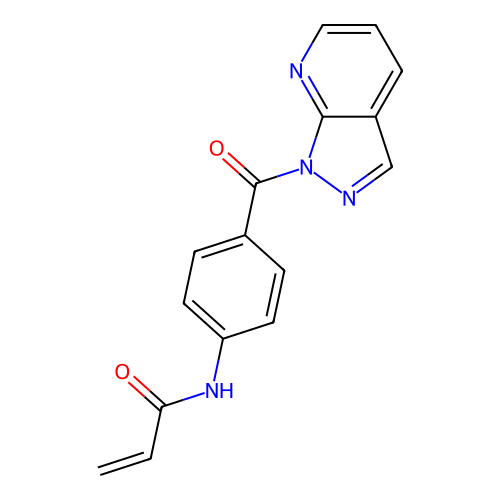
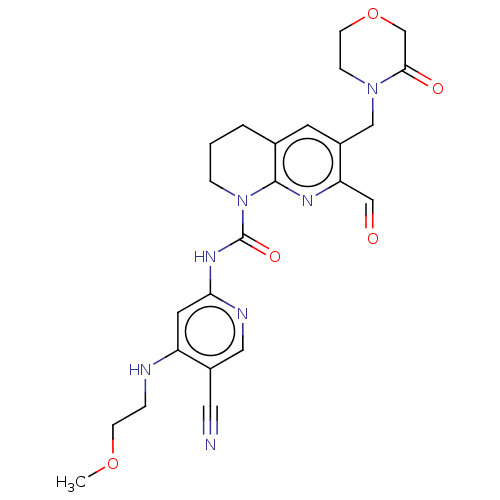
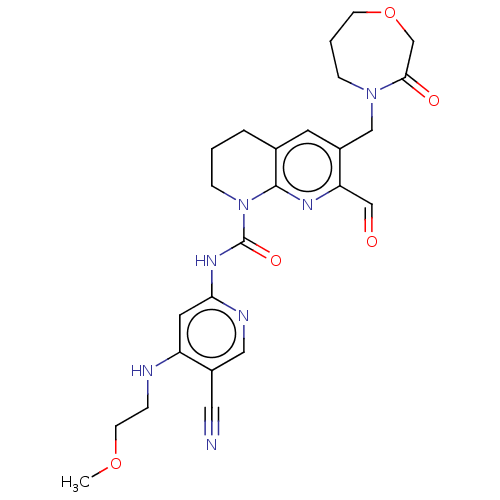
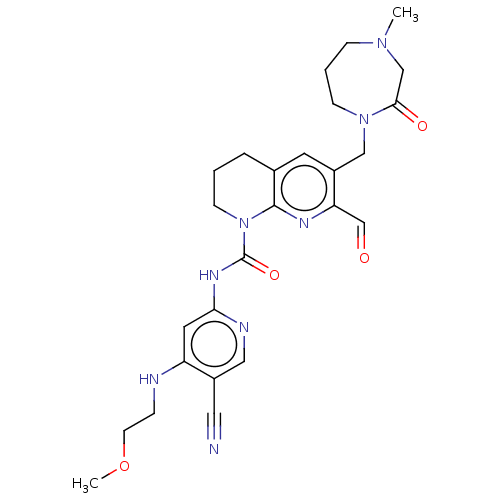
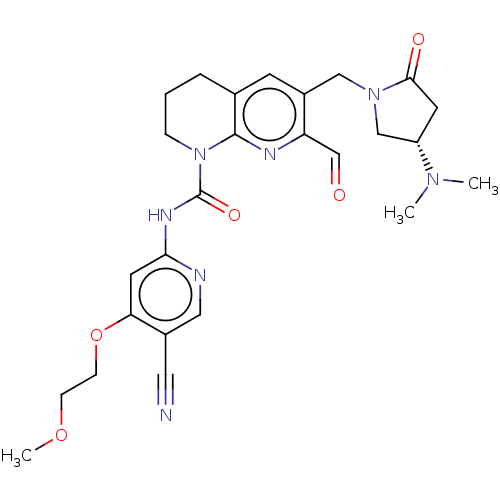
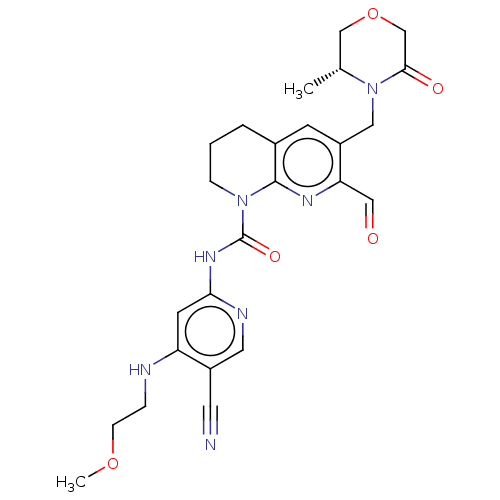
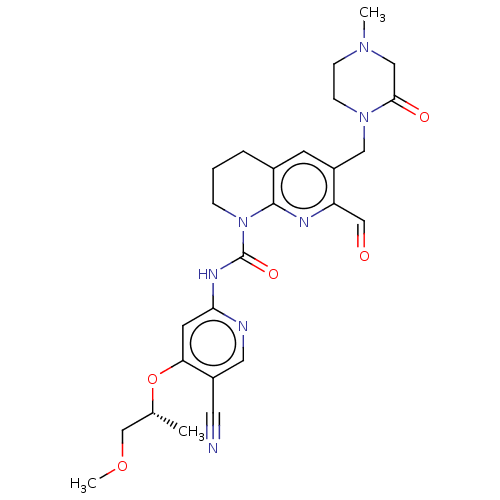
Report error Found 4757 Enz. Inhib. hit(s) with Target = 'Fibroblast growth factor receptor 4'
Affinity DataIC50: 0.100nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:Recombinant FGFR1 (2.5 nM), FGFR2 (1 nM), FGFR3 (5 nM), or FGFR4 (12 nM) (Invitrogen ) was prepared as a mixture with substrate KKKSPGEYVNIEFG (SEQ I...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMAssay Description:Recombinant FGFR1 (2.5 nM), or FGFR4 (12 nM) was prepared as a mixture with substrate KKKSPGEYVNIEFG (SEQ ID NO:1) (20 μM, FGFR1 substrate); Pol...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMAssay Description:Inhibition of recombinant non-phosphorylated FGFR4 kinase domain (442 to 753) (unknown origin) expressed in Sf9 insect cells using 5-Fluo-Ahx-KKKKEEI...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMpH: 7.5 T: 2°CAssay Description:Recombinant FGFR1 (2.5 nM), or FGFR4 (12 nM) was prepared as a mixture with substrate KKKSPGEYVNIEFG (SEQ ID NO:1) (20 μM, FGFR1 substrate); Pol...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.25nMAssay Description:Inhibition of FGFR4 V550L mutant (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of recombinant non-phosphorylated FGFR4 kinase domain (442 to 753) (unknown origin) expressed in Sf9 insect cells using 5-Fluo-Ahx-KKKKEEI...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Compound preparation: Add 45 μL of DMSO to 5 μL of stock solution at a concentration of 10 mM to prepare the solution LY2874455 at 1000 M; ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.320nMAssay Description:Inhibition of FGFR4 (unknown origin) by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:In this experiment, the inhibitory effect of the compounds on FGFR4 kinase activity was tested by a fluorescence resonance energy transfer (TR-FRET) ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:Inhibition of FGFR4 (unknown origin) by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:Inhibition of N-terminal 6x-His tagged FGFR4 catalytic domain (445-753 residues) (unknown origin) using IYGEFKKK substrate and [gamma-32P]-ATP incuba...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:Inhibition of recombinant non-phosphorylated FGFR4 kinase domain (442 to 753) (unknown origin) expressed in Sf9 insect cells using 5-Fluo-Ahx-KKKKEEI...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:Inhibition of recombinant non-phosphorylated FGFR4 kinase domain (442 to 753) (unknown origin) expressed in Sf9 insect cells using 5-Fluo-Ahx-KKKKEEI...More data for this Ligand-Target Pair
Affinity DataIC50: 0.420nMAssay Description:The FGFR4 kinase inhibition of compounds was tested by using mobility shift assay in this experiment, and the rate of FGFR4 kinase inhibition of comp...More data for this Ligand-Target Pair
Affinity DataIC50: 0.460nMAssay Description:Inhibition of FGFR4 (unknown origin) by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.470nMAssay Description:Inhibition of recombinant FGFR4 (unknown origin) by mobility assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:The inhibitory effect of the compounds on FGFR4 kinase activity was measured according to the method of Test Example 3. Purified recombinant human FG...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of N-terminal GST tagged cytoplasmic domain human FGFR4 (460 to 802 residues) expressed in Escherichia coli BL21 (DE3) using CSox as subst...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of N-terminal GST tagged cytoplasmic domain human FGFR4 (460 to 802 residues) expressed in Escherichia coli BL21 (DE3) using CSox as subst...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of N-terminal 6His-tagged FGFR4 (unknown orgin) (445 to 753 residues) expressed in Rosetta (NEB) cells using poly(Glu, Tyr) 4:1 peptide as...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Purified recombinant human FGFR4 protein was purchased from Carna Biosciences, Inc. When setting conditions for the measurement of the inhibitory ef...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of wild type FGFR4 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.550nMAssay Description:Inhibition of N-terminal GST fusion tagged human FGFR4 cytoplasmic domain (460 to 802 end residues) expressed in baculovirus expression system preinc...More data for this Ligand-Target Pair
Affinity DataIC50: 0.570nMAssay Description:Inhibition of FGFR4 (unknown origin) by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.600nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.600nMAssay Description:In this experiment, the inhibitory effect of the compounds on FGFR4 kinase activity was tested by a fluorescence resonance energy transfer (TR-FRET) ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.600nMAssay Description:Inhibition of recombinant non-phosphorylated FGFR4 kinase domain (442 to 753) (unknown origin) expressed in Sf9 insect cells using 5-Fluo-Ahx-KKKKEEI...More data for this Ligand-Target Pair
Affinity DataIC50: 0.600nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.600nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.620nMAssay Description:Inhibition of FGFR4 (unknown origin) by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.630nMAssay Description:Inhibition of FGFR4 (unknown origin) by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.630nMAssay Description:Compound preparation: Add 45 μL of DMSO to 5 μL of stock solution at a concentration of 10 mM to prepare the solution LY2874455 at 1000 M; ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.670nMAssay Description:Compound preparation: Add 45 μL of DMSO to 5 μL of stock solution at a concentration of 10 mM to prepare the solution LY2874455 at 1000 M; ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.680nMAssay Description:Inhibition of FGFR4 (unknown origin) by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.700nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.700nMAssay Description:Inhibition of FGFR4 (unknown origin) using Poly (Glu,Tyr) 4:1 as substrate incubated for 60 mins in presence of ATP by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.700nMpH: 7.5 T: 2°CAssay Description:Liquid handling and incubation steps were done on an Innovadyne Nanodrop Express equipped with a robotic arm (Thermo CatX, Caliper Twister II) and an...More data for this Ligand-Target Pair
Affinity DataIC50: 0.700nMAssay Description:Inhibition of recombinant non-phosphorylated FGFR4 kinase domain (442 to 753) (unknown origin) expressed in Sf9 insect cells using 5-Fluo-Ahx-KKKKEEI...More data for this Ligand-Target Pair