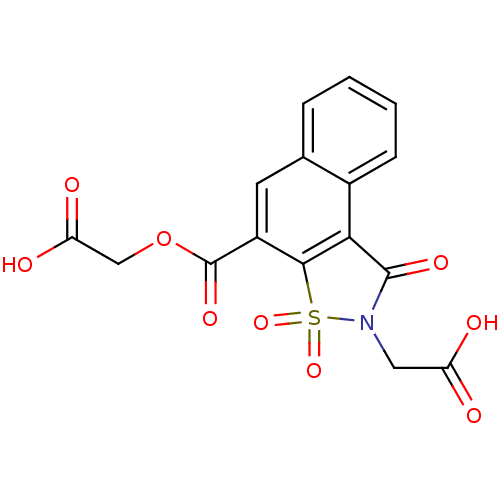
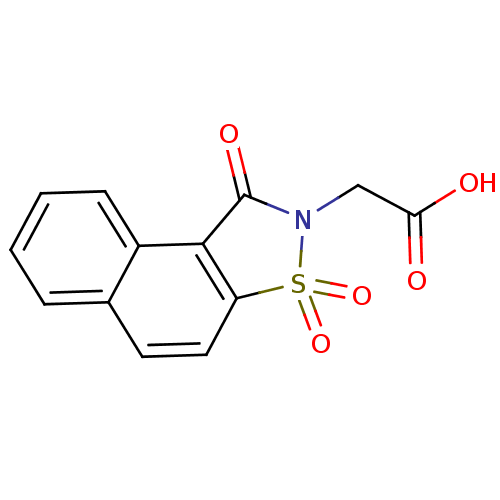
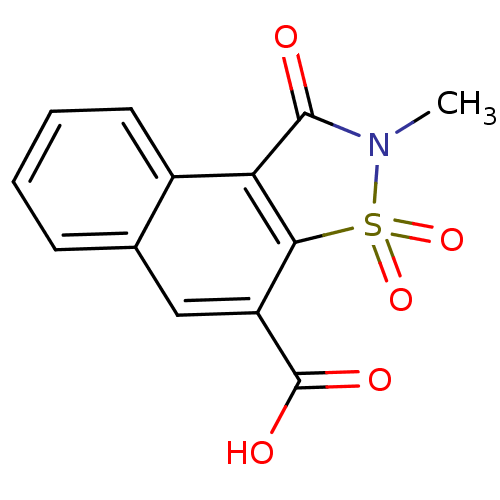
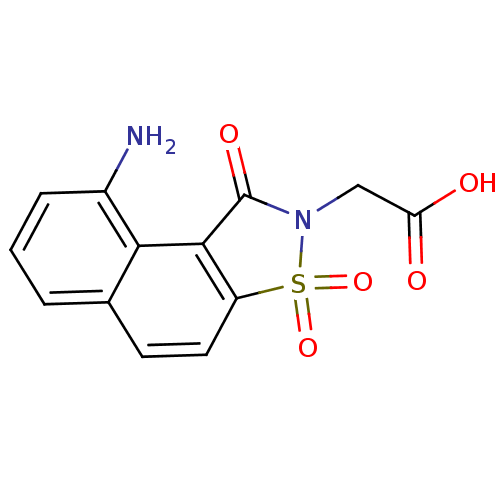
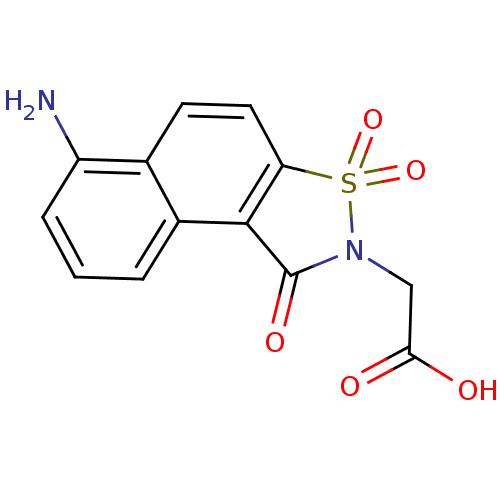
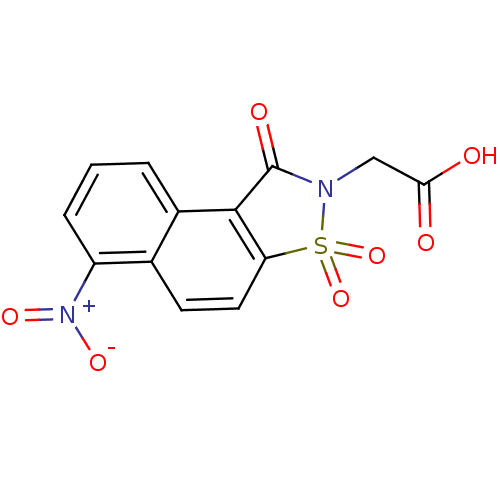
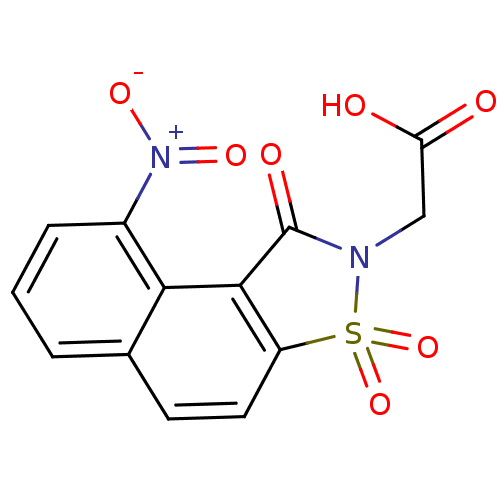
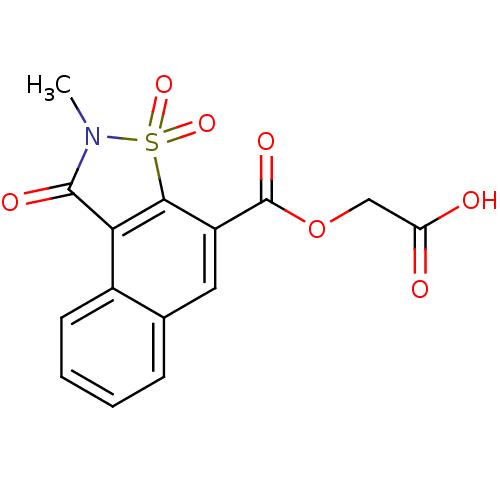
Report error Found 9 Enz. Inhib. hit(s) with all data for entry = 2032
Affinity DataIC50: 140nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DataIC50: 8.80E+4nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+5nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+5nMpH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
Affinity DatapH: 6.2 T: 2°CAssay Description:The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)