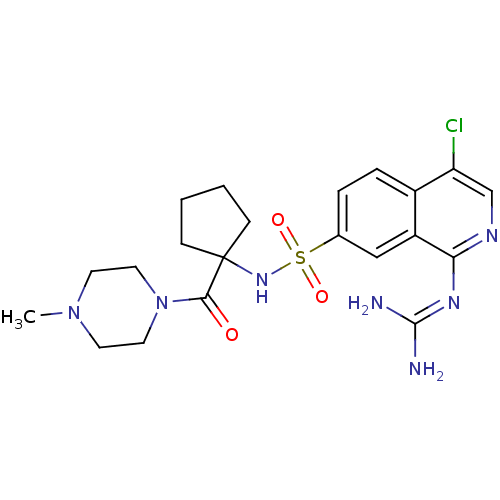
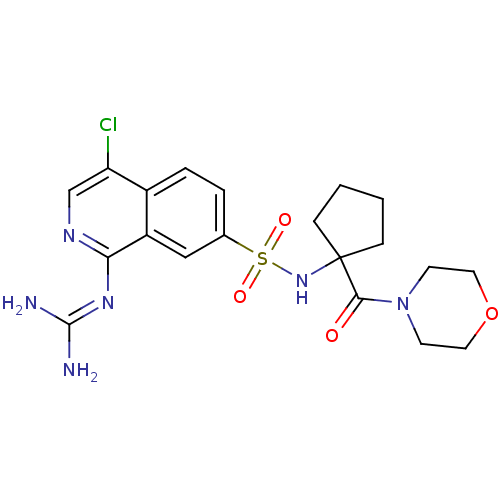
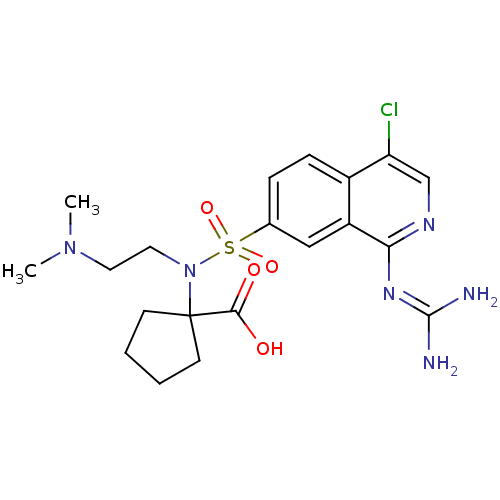
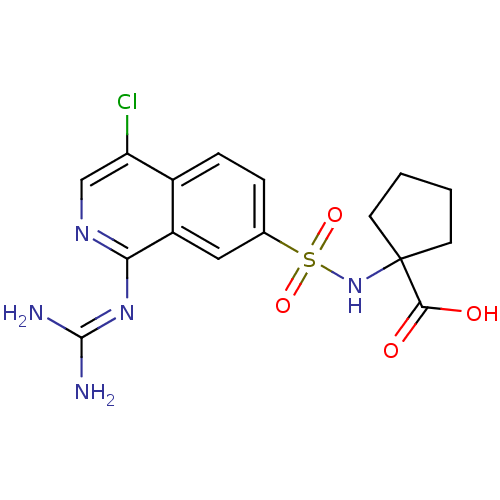
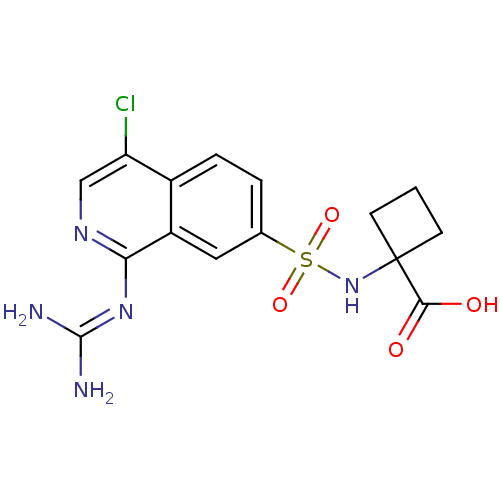
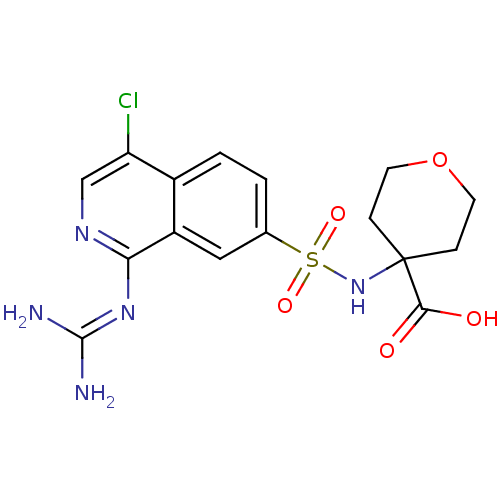
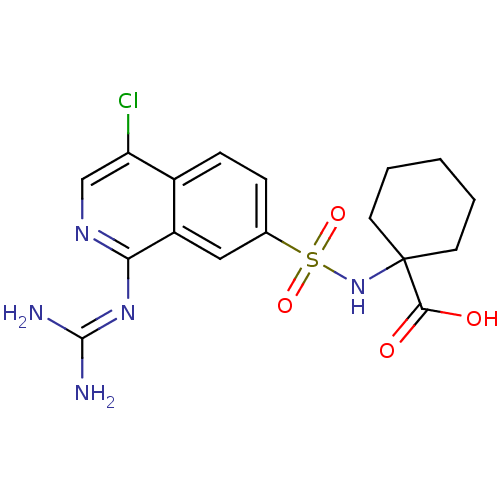
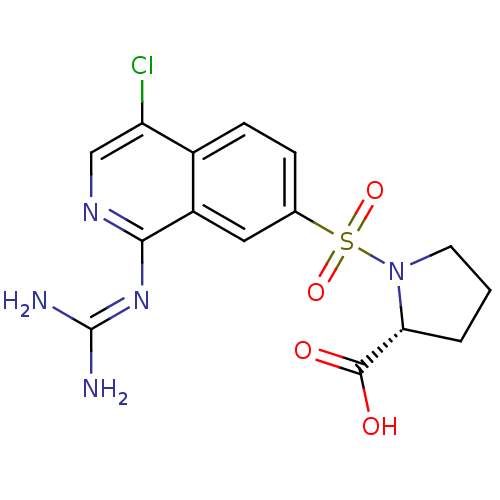
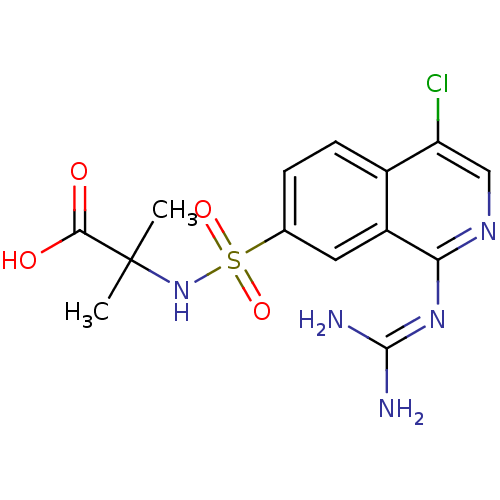
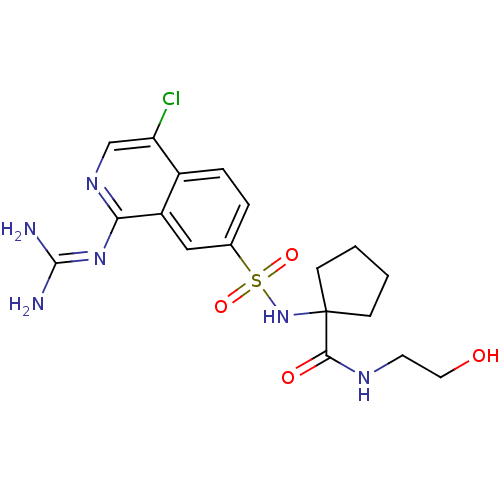
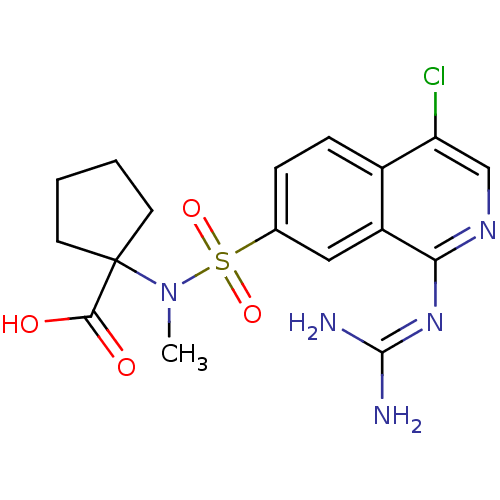
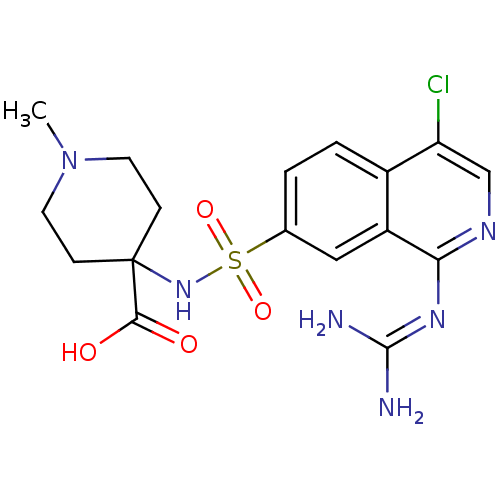
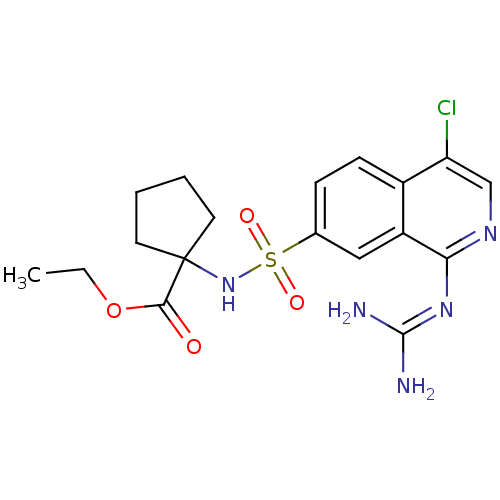
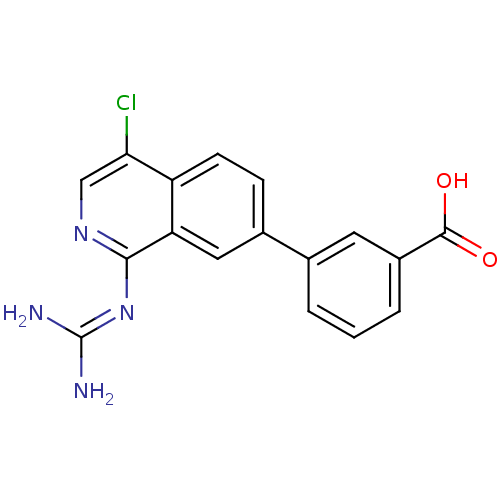
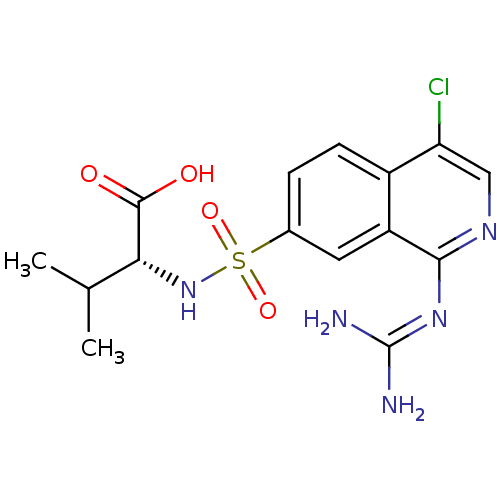
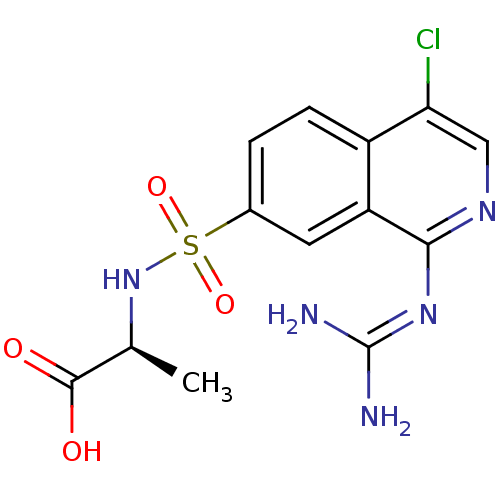
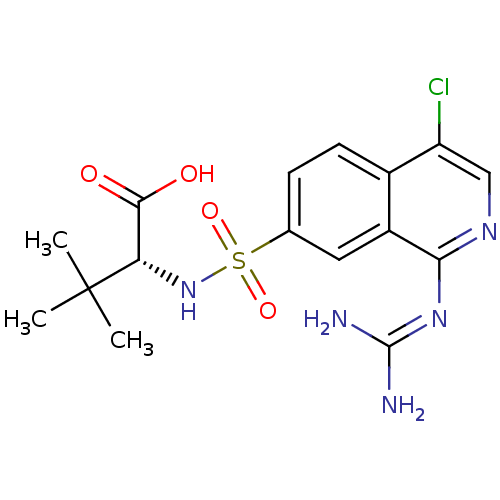
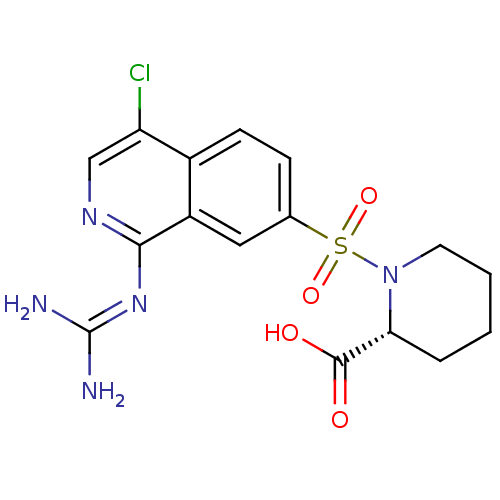
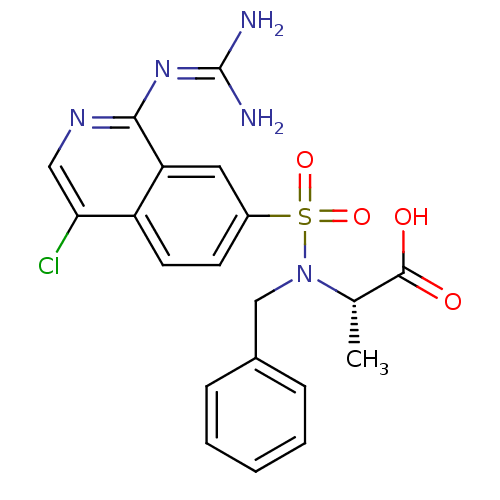
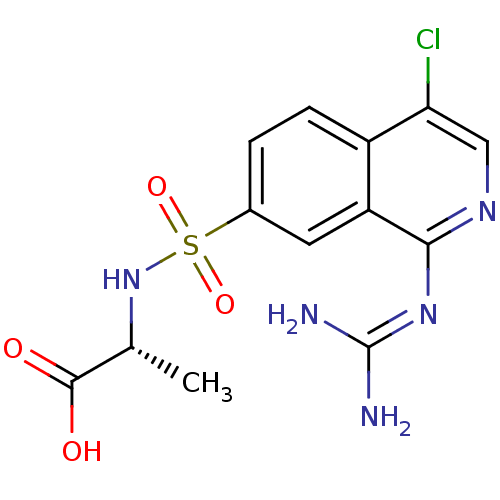
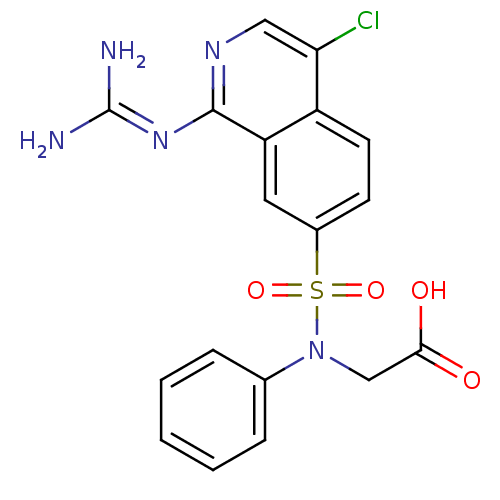
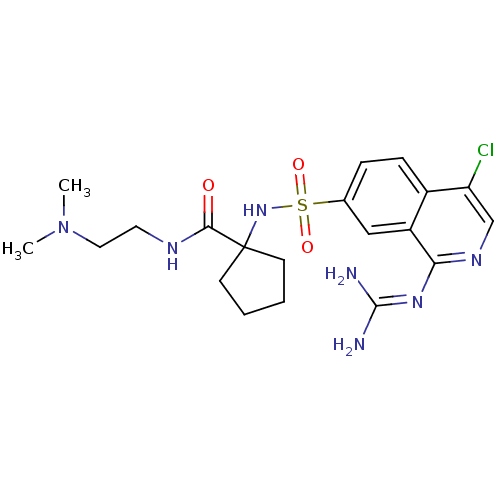
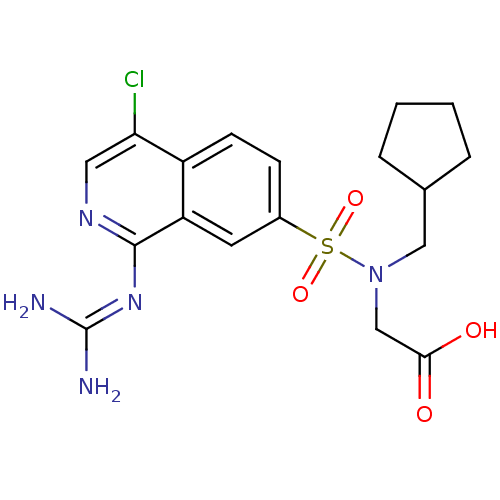
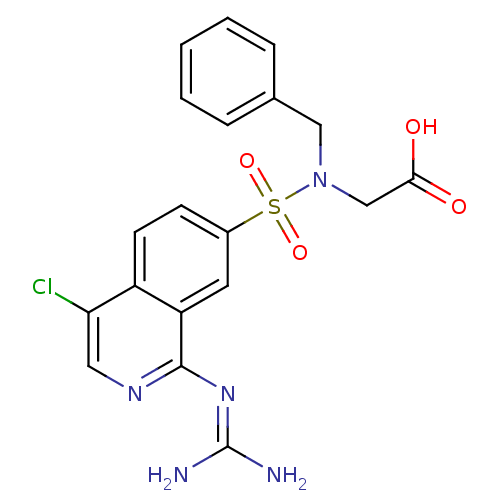
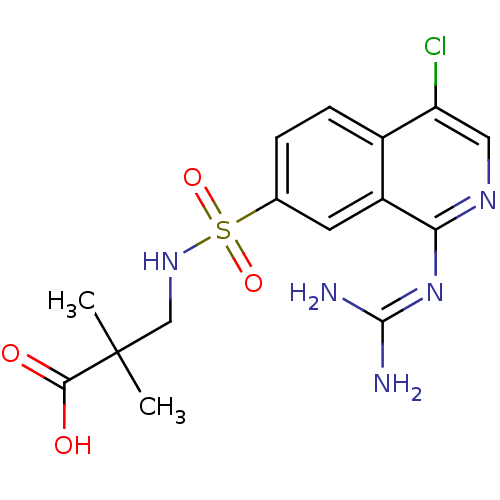
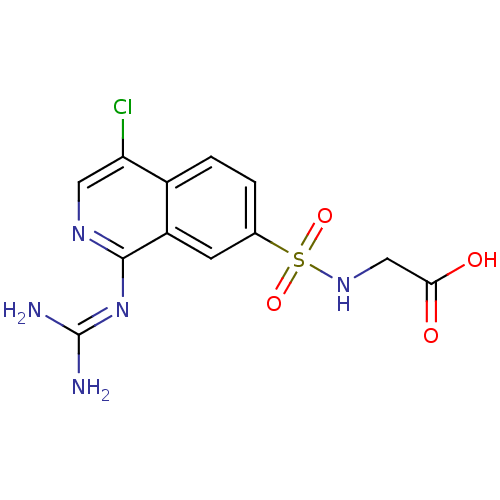
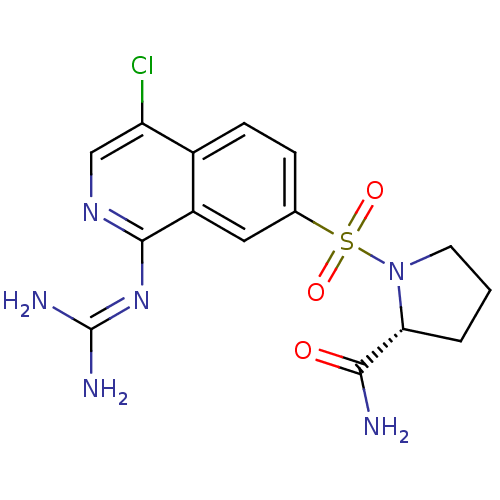
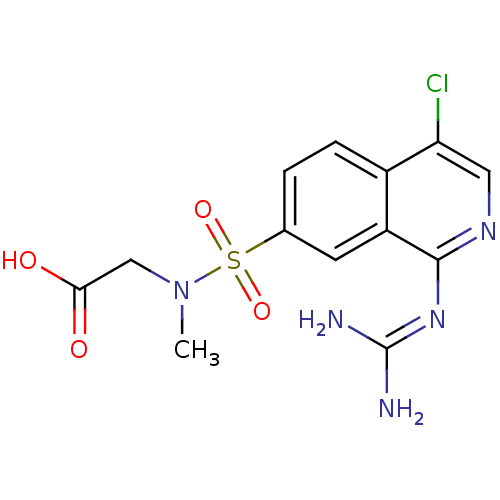
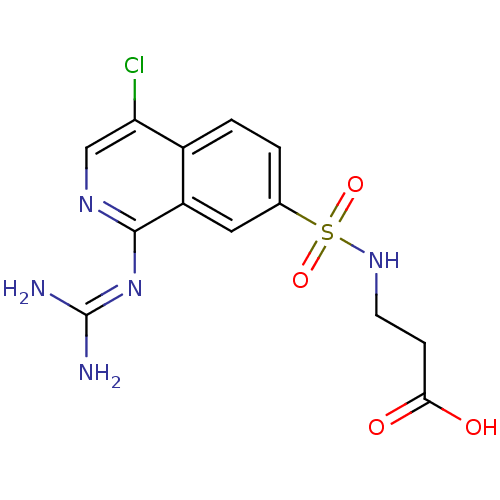
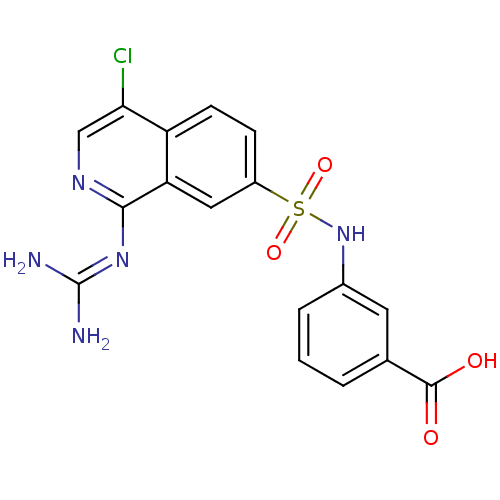
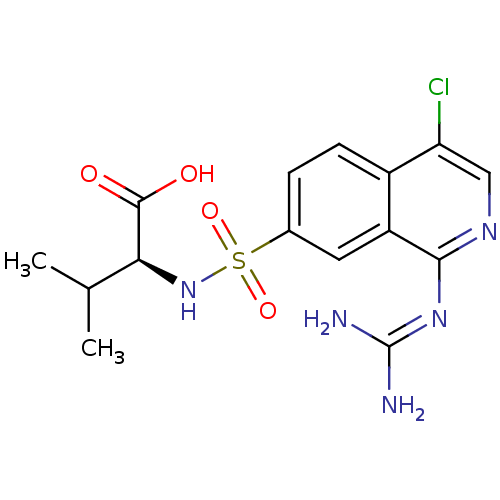
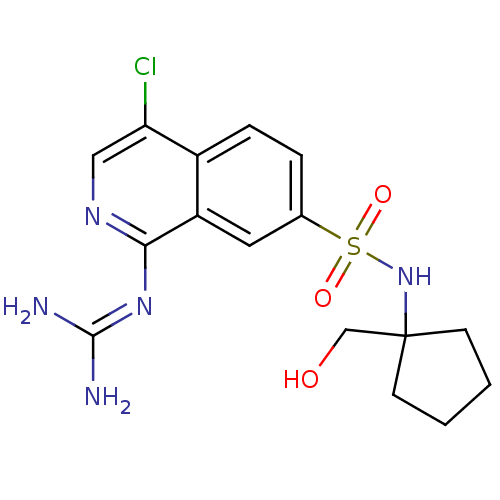
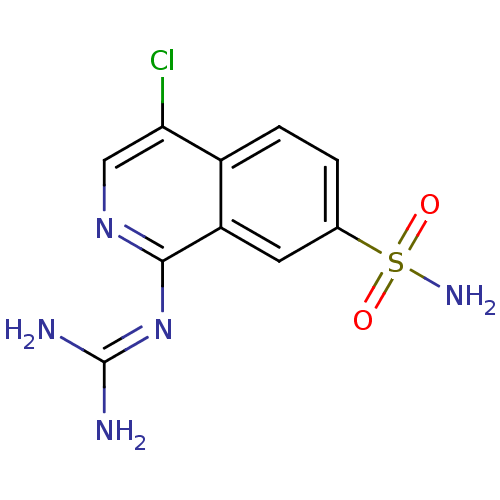
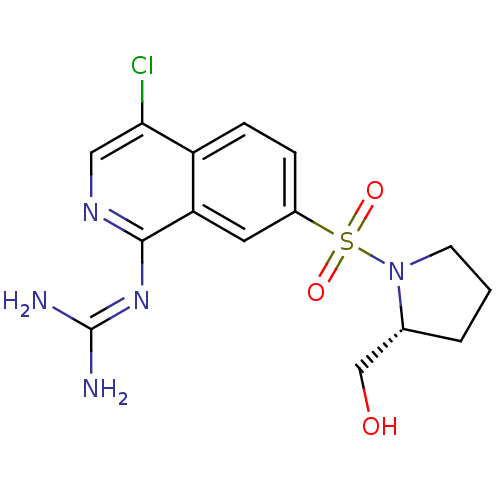
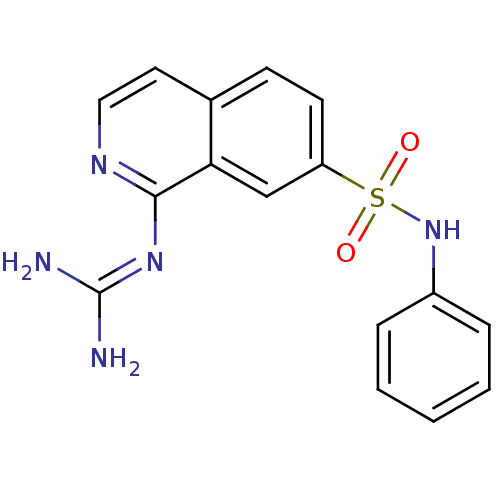
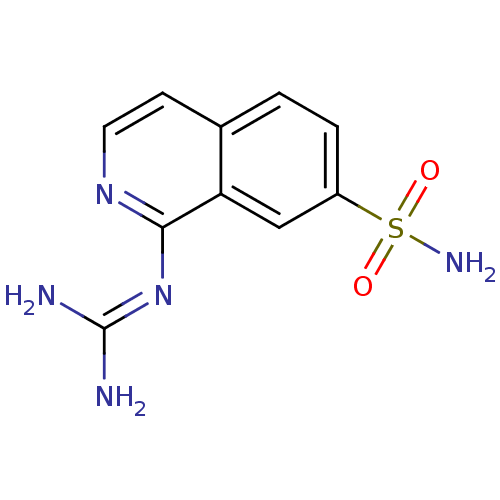
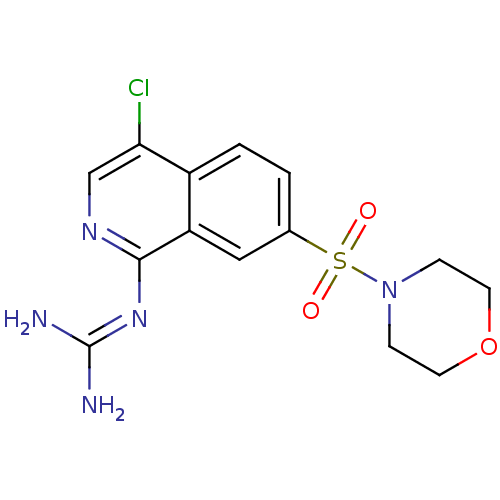
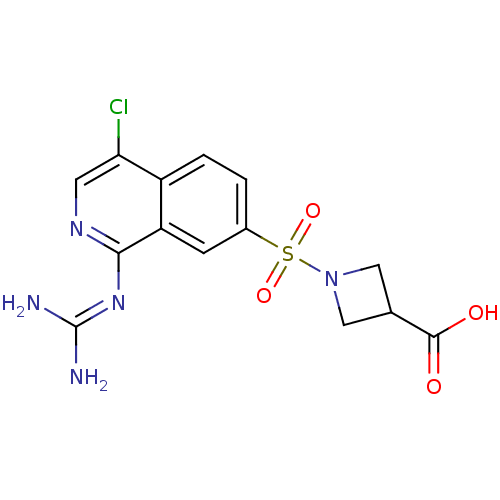
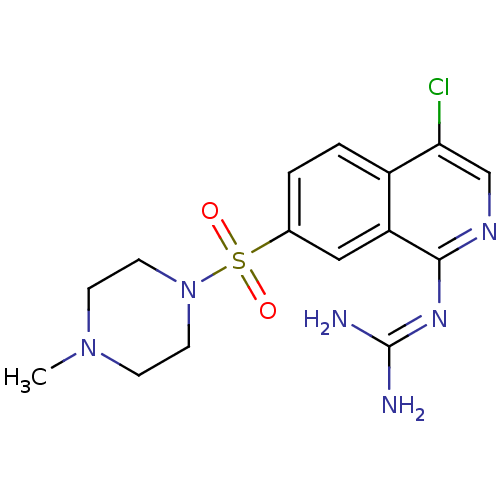
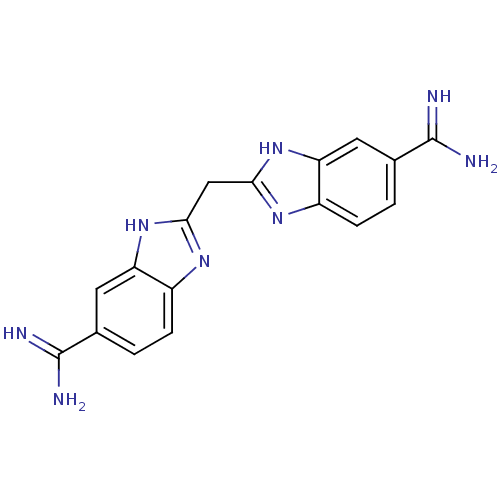
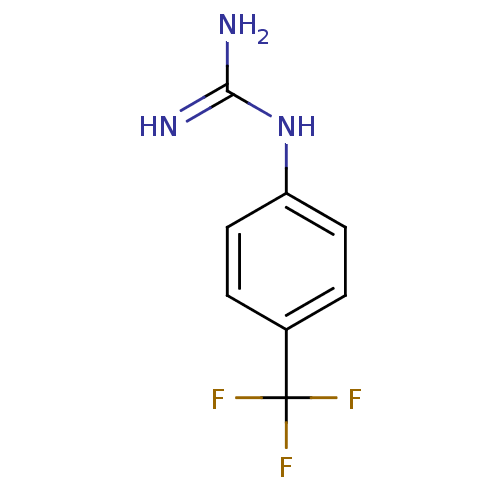
Report error Found 107 Enz. Inhib. hit(s) with all data for entry = 2017
Affinity DataKi: 1.40nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 1.60nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 2.60nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 5.80nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 7.40nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 7.40nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 8nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 9.90nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 14nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 17nM ΔG°: -46.1kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 19nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 21nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 22nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 23nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 27nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 28nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 29nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 30nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 31nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 35nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 43nM ΔG°: -43.7kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 48nM ΔG°: -43.5kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 54nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 54nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 58nM ΔG°: -43.0kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 59nM ΔG°: -42.9kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 66nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 67nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 71nM ΔG°: -42.4kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 86nM ΔG°: -42.0kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 111nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 130nM ΔG°: -40.9kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 140nM ΔG°: -40.7kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 160nM ΔG°: -40.3kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 280nM ΔG°: -38.9kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 330nM ΔG°: -38.5kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 330nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 550nM ΔG°: -37.2kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 2.30E+3nM ΔG°: -33.5kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 3.20E+3nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 6.00E+3nM ΔG°: -31.0kJ/molepH: 8.1 T: 2°CAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 9.80E+3nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 1.30E+4nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 1.40E+4nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair
Affinity DataKi: 1.50E+4nMAssay Description:Ki values for compounds were calculated by incubation of each enzyme with its substrate and various compound concentrations. Absorbance was read at 4...More data for this Ligand-Target Pair