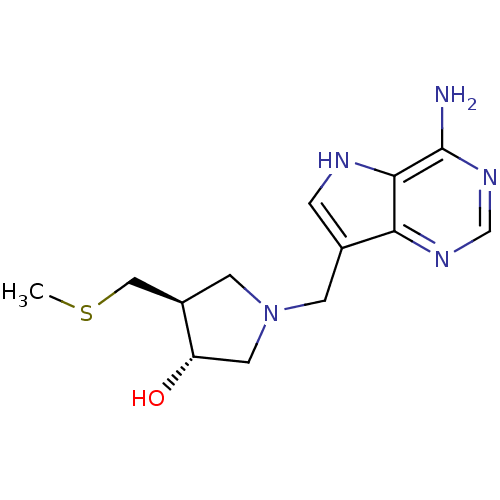
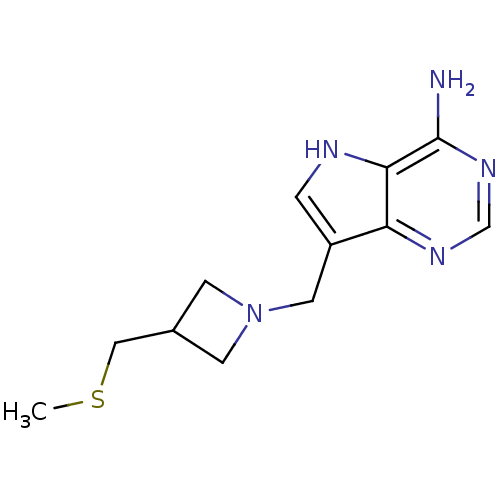
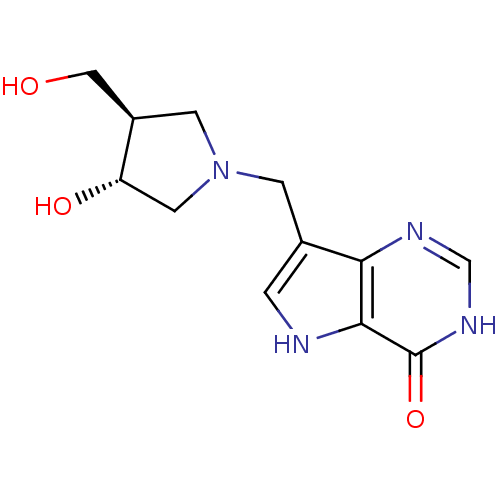
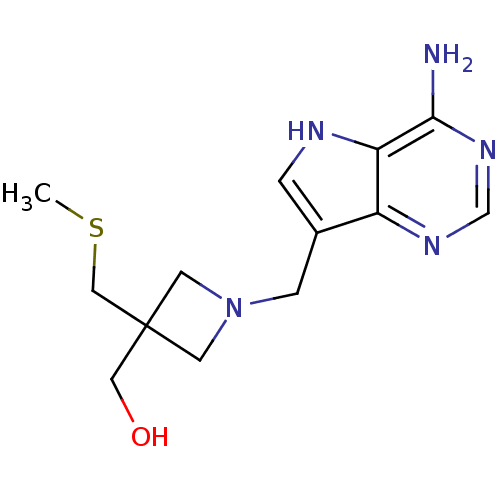
Report error Found 27 Enz. Inhib. hit(s) with all data for entry = 2567
Target5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase(Escherichia coli (strain K12))
Industrial Research
Industrial Research
Affinity DataKi: 0.0480nM ΔG°: -58.3kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 0.229nM ΔG°: -54.5kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Target5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase(Escherichia coli (strain K12))
Industrial Research
Industrial Research
Affinity DataKi: 0.450nM ΔG°: -52.8kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 0.5nM ΔG°: -52.6kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 0.665nM ΔG°: -51.9kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Target5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase(Escherichia coli (strain K12))
Industrial Research
Industrial Research
Affinity DataKi: 0.840nM ΔG°: -51.3kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 1nM ΔG°: -50.9kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 1.10nM ΔG°: -50.6kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 1.70nM ΔG°: -49.6kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 1.80nM ΔG°: -49.4kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 1.80nM ΔG°: -49.4kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 2nM ΔG°: -49.2kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 4.80nM ΔG°: -47.0kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 6.30nM ΔG°: -46.3kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 12.9nM ΔG°: -44.6kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 16nM ΔG°: -44.0kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 24nM ΔG°: -43.1kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 84nM ΔG°: -40.0kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 140nM ΔG°: -38.7kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 150nM ΔG°: -38.6kJ/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 191nM ΔG°: -38.0kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 280nM ΔG°: -37.0kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 360nM ΔG°: -36.4kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 580nM ΔG°: -35.2kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: 1.29E+3nM ΔG°: -33.3kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nM ΔG°: >-28.3kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nM ΔG°: >-28.3kJ/molepH: 7.7 T: 2°CAssay Description:PNP activity was monitored by absorbance change in a coupled assay. In the assay, inosine was converted to hypoxanthine, and then hypoxanthine was co...More data for this Ligand-Target Pair