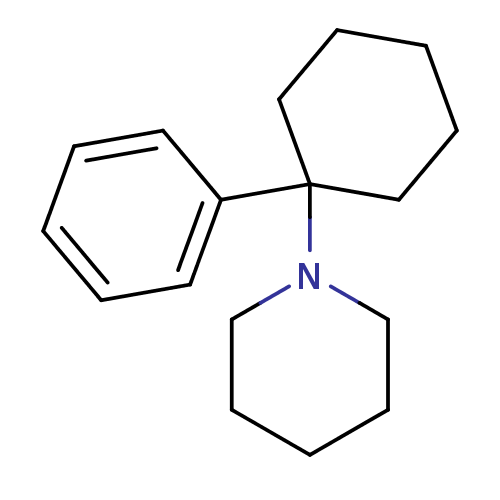
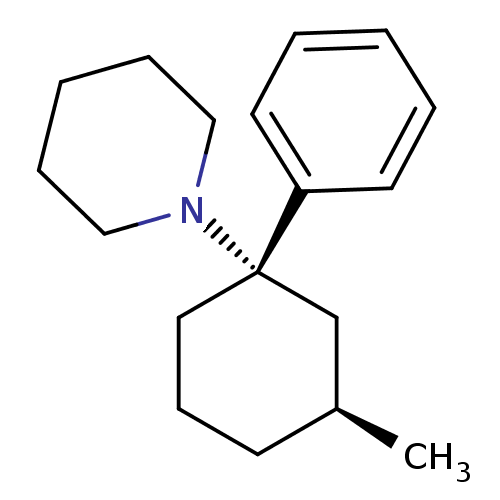
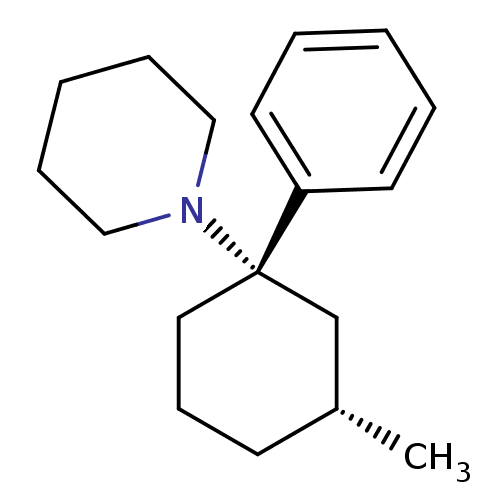
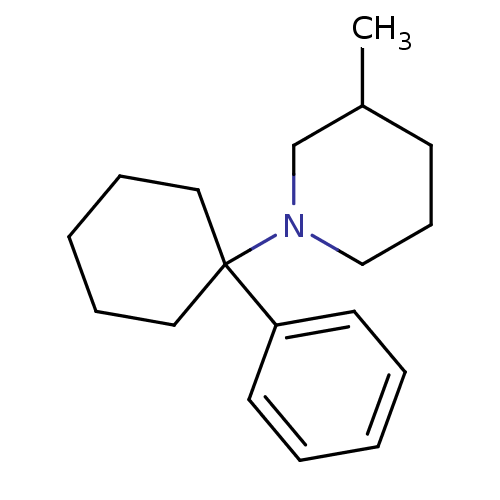
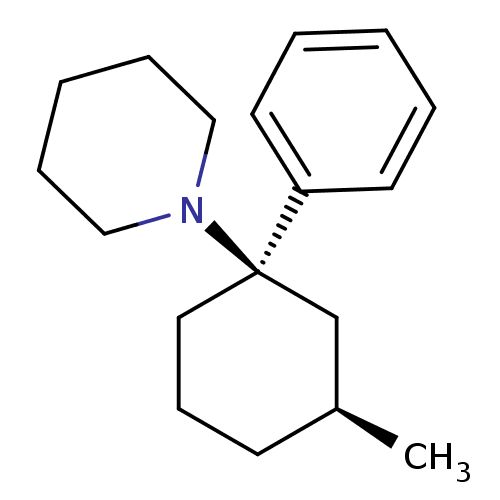
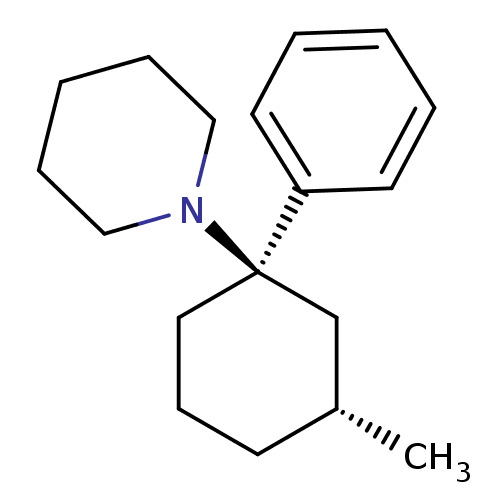
Report error Found 8 Enz. Inhib. hit(s) with all data for entry = 50001942
Affinity DataKi: 60nMAssay Description:Binding affinity against phencyclidine receptor was determined by the displacement of [3H]TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair
Affinity DataKi: 170nMAssay Description:Binding affinity against phencyclidine receptor was determined by the displacement of [3H]TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair
Affinity DataIC50: 600nMAssay Description:Inhibition activity against binding of phencyclidine receptor was determined by the displacement of [3H]-TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair
Affinity DataKi: 820nMAssay Description:Binding affinity against phencyclidine receptor was determined by the displacement of [3H]TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair
Affinity DataKi: 2.15E+3nMAssay Description:Binding affinity against phencyclidine receptor was determined by the displacement of [3H]TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair
Affinity DataKi: 2.15E+3nMAssay Description:Binding affinity against phencyclidine receptor was determined by the displacement of [3H]TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair
Affinity DataKi: 3.30E+3nMAssay Description:Binding affinity against phencyclidine receptor was determined by the displacement of [3H]TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair
Affinity DataKi: 6.80E+3nMAssay Description:Binding affinity against phencyclidine receptor was determined by the displacement of [3H]TCP of whole rat brain minus cerebellum.More data for this Ligand-Target Pair