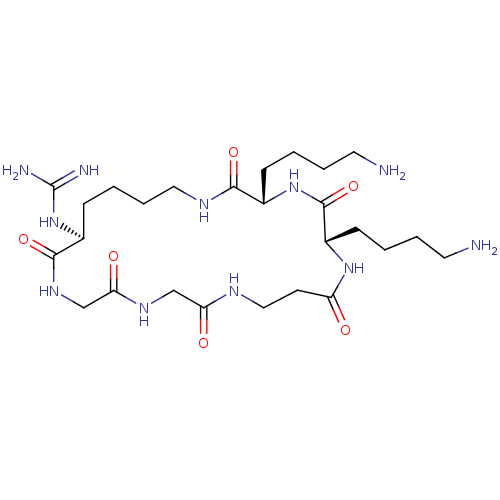
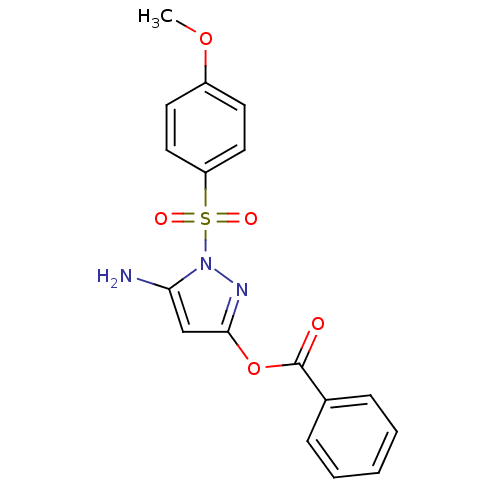
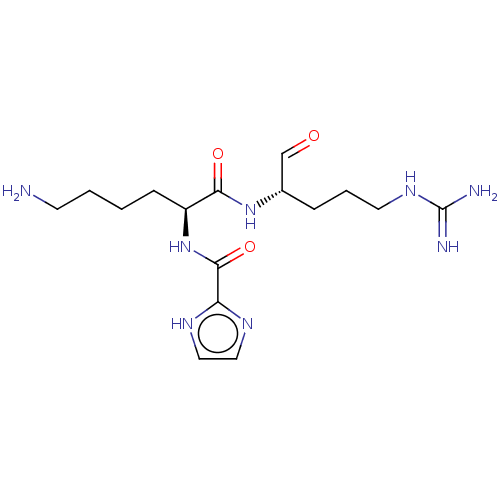
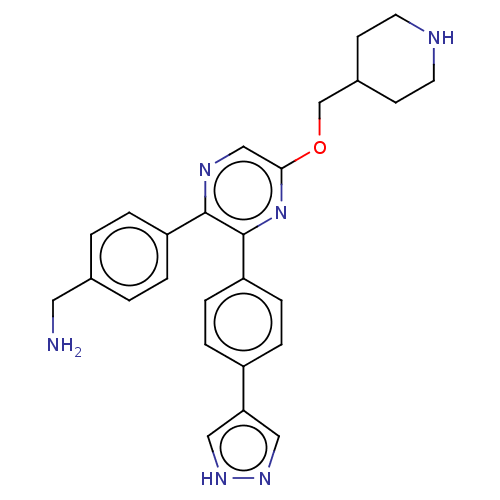
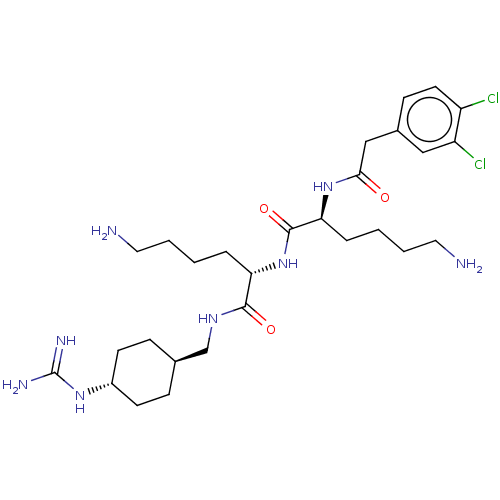
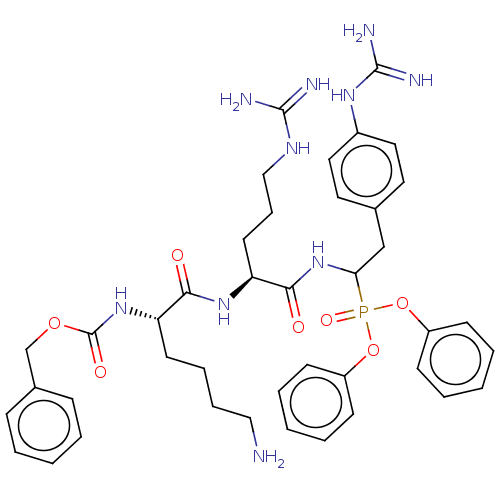
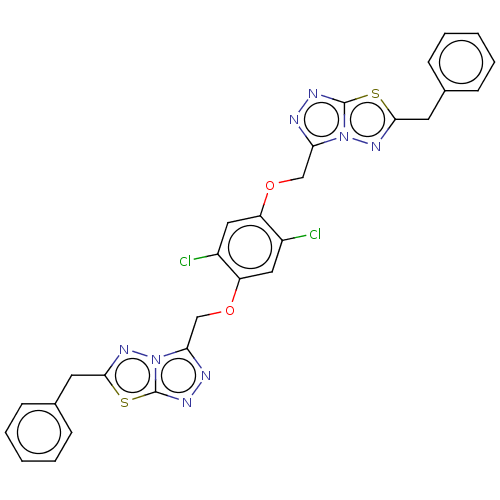
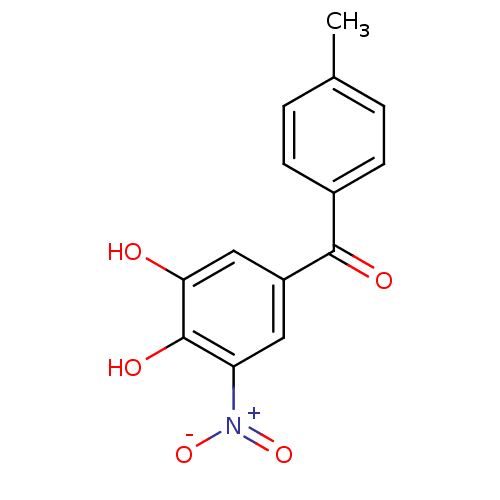
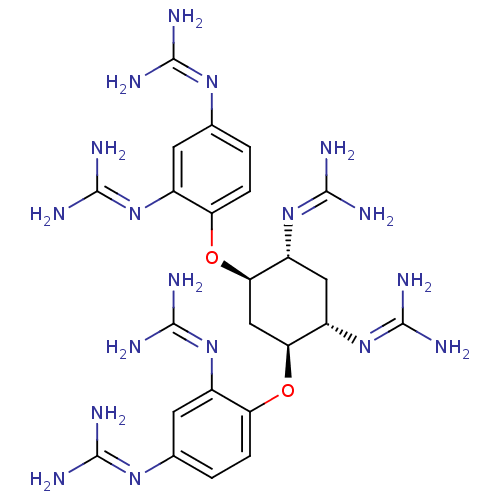
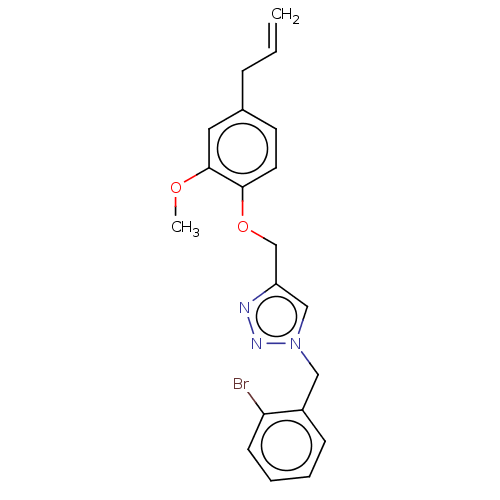
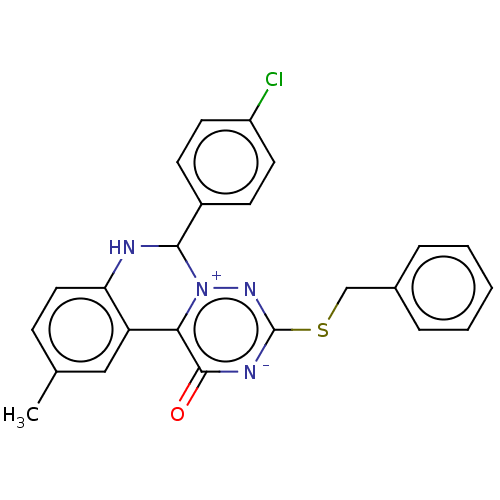
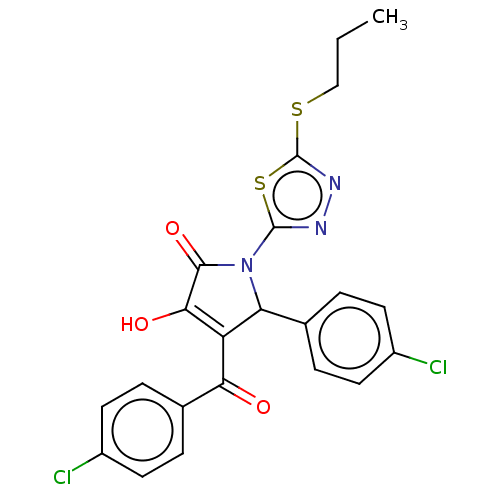
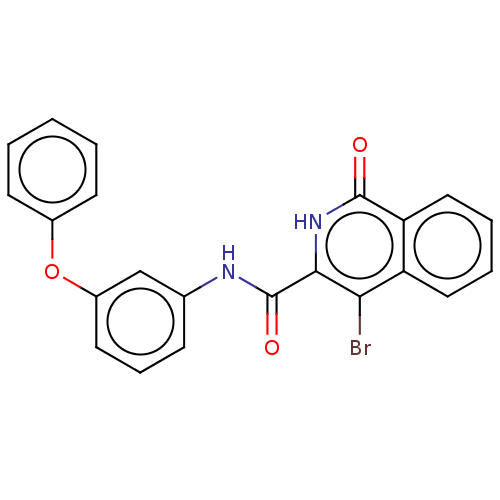
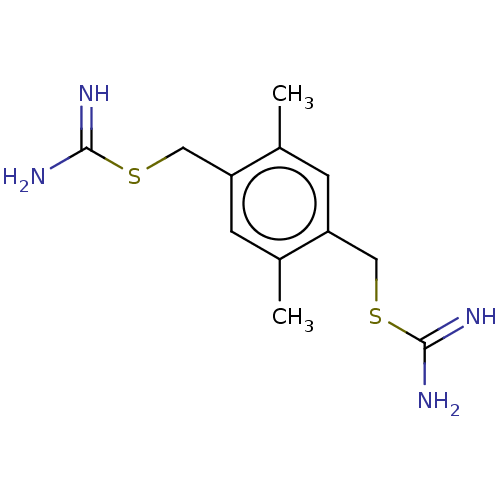
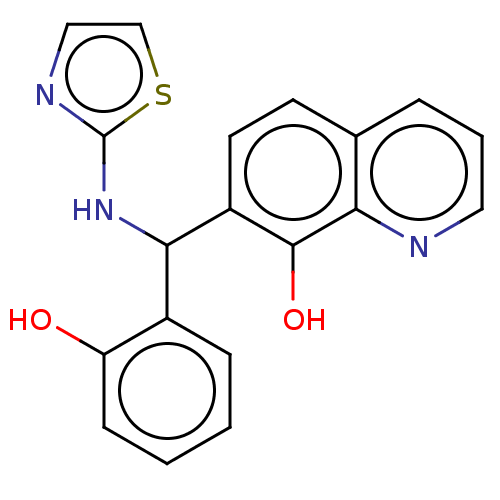
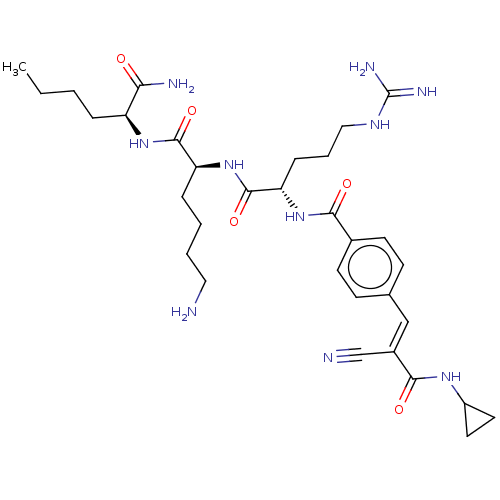
Report error Found 24 Enz. Inhib. hit(s) with all data for entry = 50004309
Affinity DataKi: 1.70nMAssay Description:Inhibition of Zika virus NS2B-NS3 protease using PhAc-Leu-LysLys-Arg-AMC as substrate measured for 10 mins by fluorescence plate reader analysisMore data for this Ligand-Target Pair
Affinity DataKi: 69nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease using PhAc-Leu-LysLys-Arg-AMC as substrate measured for 10 mins by fluorescence plate reader analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 105nMAssay Description:Inhibition of West Nile virus NS2B (1393 to 1440 residues)-NS3 (1476 to 1687 residues) protease expressed in Escherichia coli BL21 (DE3) codon plus c...More data for this Ligand-Target Pair
Affinity DataIC50: 120nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease expressed in Escherichia coli BL21 (DE3) RILP codon plus competent cells using Benzoyl-NleKRR-AMC as ...More data for this Ligand-Target Pair
Affinity DataIC50: 120nMAssay Description:Inhibition of West Nile virus NS2B (49 to 96 residues)-NS3 (2 to 184 residues) protease expressed in Escherichia coli BL21-rosetta cells using Bz-Nle...More data for this Ligand-Target Pair
Affinity DataKi: 130nMAssay Description:Inhibition of West Nile virus NS2B (52 to 96 residues)-NS3 (1 to 184 residues) protease using PhAc-Leu-LysLys-Arg-AMC3 TFA as substrate by fluorescen...More data for this Ligand-Target Pair
Affinity DataKi: 400nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease using Pyr-RTKR-AMC as substrate preincubated for 10 mins followed by substrate addition and measured ...More data for this Ligand-Target Pair
Affinity DataIC50: 440nMAssay Description:Inhibition of 6his tagged West Nile virus NS2B-NS3 protease expressed in Escherichia coli BL21 CodonPlus (DE3)-RIPL cells using Pyr-RTKR-AMC as subst...More data for this Ligand-Target Pair
Affinity DataIC50: 700nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease expressed in Escherichia coli using Bz-Nle-Lys-Arg-Arg-AMC as substrate assessed as proteolytic activ...More data for this Ligand-Target Pair
Affinity DataIC50: 710nMAssay Description:Inhibition of Zika virus NS2B (47 to 95 residues)-NS3 (1 to 170 residues) protease expressed in Escherichia coli BL21-rosetta cells using Bz-Nle-Lys-...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+3nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease expressed in Escherichia coli using Phe-Ala-Ser-Gly-Lys-Arg-pNA assessed as proteolytic cleavage of t...More data for this Ligand-Target Pair
Affinity DataKi: 3.06E+3nMAssay Description:Inhibition of recombinant West Nile virus NS2B-NS3 protease using pERTKR-AMC as substrate assessed as inhibition constant by fluorescence based assayMore data for this Ligand-Target Pair
Affinity DataKi: 4.47E+3nMAssay Description:Uncompetitive inhibition of recombinant West Nile virus NS2B-NS3 protease using Pyr-RTKR-AMC as substrate assessed as inhibition constant preincubate...More data for this Ligand-Target Pair
Affinity DataIC50: 6.10E+3nMAssay Description:Inhibition of recombinant West Nile virus NS2B-NS3 protease using Pyr-RTKR-AMC as substrate assessed as change in fluorescence intensity preincubated...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease using Boc-Gly-Lys-Arg-7-amino-4-methyl coumarin as substrate preincubated for 15 mins followed by sub...More data for this Ligand-Target Pair
Affinity DataKd: 4.00E+4nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease assessed as dissociation constant by NMR spectroscopic anlaysisMore data for this Ligand-Target Pair
TargetPlatelet-derived growth factor receptor alpha/beta(Mouse)
Australian National University Canberra
Curated by ChEMBL
Australian National University Canberra
Curated by ChEMBL
Affinity DataIC50: 5.00E+4nMAssay Description:Inhibition of trypsin (unknown origin) using enzyloxy-carbonyl (Z)-Gly-Pro-Arg-para-nitroanilide as substrate preincubated with compound for 15 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+4nMAssay Description:Inhibition of factor Xa (unknown origin) using Boc-Ile-Glu-Gly-Arg-AMC as substrate preincubated with compound for 15 mins followed by substrate addi...More data for this Ligand-Target Pair
Affinity DataKi: 9.25E+4nMAssay Description:Mixed type inhibition of West Nile virus NSB2-NS3 protease using Abz-Nle-Lys-Arg-Arg-Ser-3-(NO2)Tyr as substrate preincubated with compound for 15 mi...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of furin (unknown origin) using Pyr-RTKR-AMC as substrateMore data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+5nMAssay Description:Inhibition of West Nile virus NS2B-NS3 proteaseMore data for this Ligand-Target Pair
Affinity DataKi: >2.00E+5nMAssay Description:Inhibition of factor Xa (unknown origin) using CH3OCO-dCha-Gly-Arg-pNA as substrate by fluorescence plate reader analysisMore data for this Ligand-Target Pair
Affinity DataKi: 5.00E+5nMAssay Description:Inhibition of West Nile virus NS2B-NS3 protease expressed in Escherichia coli BL21 (DE3) cells using Bz-Nle-K-K-R-AM as substrate preincubated for 10...More data for this Ligand-Target Pair
Affinity DataKi: >5.00E+5nMAssay Description:Inhibition of thrombin (unknown origin) using CH3SO2-dCha-Gly-Arg-pNA as substrate by fluorescence plate reader analysisMore data for this Ligand-Target Pair